Co-evolution within structured bacterial communities results in multiple expansion of CRISPR loci and enhanced immunity
- PMID: 32223887
- PMCID: PMC7105378
- DOI: 10.7554/eLife.53078
Co-evolution within structured bacterial communities results in multiple expansion of CRISPR loci and enhanced immunity
Abstract
Type II CRISPR-Cas systems provide immunity against phages and plasmids that infect bacteria through the insertion of a short sequence from the invader's genome, known as the 'spacer', into the CRISPR locus. Spacers are transcribed into guide RNAs that direct the Cas9 nuclease to its target on the invader. In liquid cultures, most bacteria acquire a single spacer. Multiple spacer integration is a rare event which significance for immunity is poorly understood. Here, we found that when phage infections occur on solid media, a high proportion of the surviving colonies display complex morphologies that contain cells with multiple spacers. This is the result of the viral-host co-evolution, in which the immunity provided by the initial acquired spacer is easily overcome by escaper phages. Our results reveal the versatility of CRISPR-Cas immunity, which can respond with both single or multiple spacer acquisition schemes to solve challenges presented by different environments.
Keywords: CRISPR; bacteriophage; coevolution; infectious disease; microbiology.
© 2020, Pyenson and Marraffini.
Conflict of interest statement
NP No competing interests declared, LM is a cofounder and Scientific Advisory Board member of Intellia Therapeutics, and a co-founder of Eligo Biosciences.
Figures


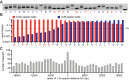




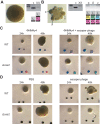


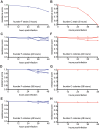

Comment in
-
An adaptable defense.Elife. 2020 Mar 30;9:e56122. doi: 10.7554/eLife.56122. Elife. 2020. PMID: 32223888 Free PMC article.
References
-
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. - DOI - PMC - PubMed
-
- Carte J, Christopher RT, Smith JT, Olson S, Barrangou R, Moineau S, Glover CV, Graveley BR, Terns RM, Terns MP. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Molecular Microbiology. 2014;93:98–112. doi: 10.1111/mmi.12644. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

