Bioinformatics services for analyzing massive genomic datasets
- PMID: 32224841
- PMCID: PMC7120352
- DOI: 10.5808/GI.2020.18.1.e8
Bioinformatics services for analyzing massive genomic datasets
Abstract
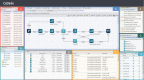
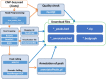
The explosive growth of next-generation sequencing data has resulted in ultra-large-scale datasets and ensuing computational problems. In Korea, the amount of genomic data has been increasing rapidly in the recent years. Leveraging these big data requires researchers to use large-scale computational resources and analysis pipelines. A promising solution for addressing this computational challenge is cloud computing, where CPUs, memory, storage, and programs are accessible in the form of virtual machines. Here, we present a cloud computing-based system, Bio-Express, that provides user-friendly, cost-effective analysis of massive genomic datasets. Bio-Express is loaded with predefined multi-omics data analysis pipelines, which are divided into genome, transcriptome, epigenome, and metagenome pipelines. Users can employ predefined pipelines or create a new pipeline for analyzing their own omics data. We also developed several web-based services for facilitating downstream analysis of genome data. Bio-Express web service is freely available at https://www.bioexpress.re.kr/.
Keywords: analysis pipeline; cloud computing; genomic data; web server; workflow system.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
