Structure-Based Design of Antivirals against Envelope Glycoprotein of Dengue Virus
- PMID: 32225021
- PMCID: PMC7232406
- DOI: 10.3390/v12040367
Structure-Based Design of Antivirals against Envelope Glycoprotein of Dengue Virus
Abstract
Dengue virus (DENV) presents a significant threat to global public health with more than 500,000 hospitalizations and 25,000 deaths annually. Currently, there is no clinically approved antiviral drug to treat DENV infection. The envelope (E) glycoprotein of DENV is a promising target for drug discovery as the E protein is important for viral attachment and fusion. Understanding the structure and function of DENV E protein has led to the exploration of structure-based drug discovery of antiviral compounds and peptides against DENV infections. This review summarizes the structural information of the DENV E protein with regards to DENV attachment and fusion. The information enables the development of antiviral agents through structure-based approaches. In addition, this review compares the potency of antivirals targeting the E protein with the antivirals targeting DENV multifunctional enzymes, repurposed drugs and clinically approved antiviral drugs. None of the current DENV antiviral candidates possess potency similar to the approved antiviral drugs which indicates that more efforts and resources must be invested before an effective DENV drug materializes.
Keywords: antiviral; dengue virus; envelope glycoprotein; structural biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

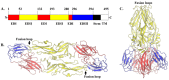
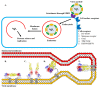

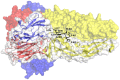
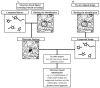

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

