Applications of heteronuclear NMR spectroscopy in biological and medicinal inorganic chemistry
- PMID: 32226090
- PMCID: PMC7094630
- DOI: 10.1016/j.ccr.2008.01.016
Applications of heteronuclear NMR spectroscopy in biological and medicinal inorganic chemistry
Abstract
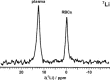
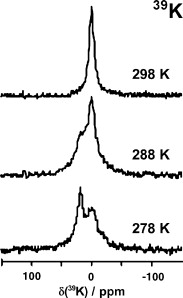
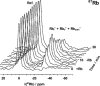
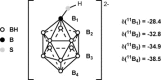
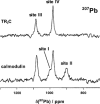
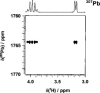
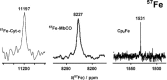
There is a wide range of potential applications of inorganic compounds, and metal coordination complexes in particular, in medicine but progress is hampered by a lack of methods to study their speciation. The biological activity of metal complexes is determined by the metal itself, its oxidation state, the types and number of coordinated ligands and their strength of binding, the geometry of the complex, redox potential and ligand exchange rates. For organic drugs a variety of readily observed spin I = 1/2 nuclei can be used (1H, 13C, 15N, 19F, 31P), but only a few metals fall into this category. Most are quadrupolar nuclei giving rise to broad lines with low detection sensitivity (for biological systems). However we show that, in some cases, heteronuclear NMR studies can provide new insights into the biological and medicinal chemistry of a range of elements and these data will stimulate further advances in this area.
Keywords: ADP, adenosine diphosphate; AES, atomic emission spectroscopy; AMP, adenosine monophosphate; ATP, adenosine triphosphate; BNCT, boron neutron capture therapy; BPG, 2,3-bisphosphoglycerate; BSA, bovine serum albumin; BSH, sodium borocaptate; Bioinorganic chemistry; Biological systems; DNA, deoxyribonucleic acid; EDTA-N4, ethylenediaminetetraacetamide; EFG, electric field gradient; GMP, guanosine monophosphate; HMQC, heteronuclear multiple quantum correlation; Heteronuclear NMR spectroscopy; Im, imidazole; In, indazole; MQF, multiple quantum filtered; MRI, magnetic resonance imaging; Medicinal inorganic chemistry; Metallopharmaceuticals; NOE, nuclear Overhauser effect; PET, positron emission tomography; Quadrupolar nuclei; RBC, red blood cell; RNA, ribonucleic acid; SDS, sodium dodecyl sulfate; rRNA, ribosomal ribonucleic acid; tRNA, transfer ribonucleic acid.
Copyright © 2008 Elsevier B.V. All rights reserved.
Figures














References
-
- McMaster J. Annu. Rep. Prog. Chem. Sect. A: Inorg. Chem. 2002;98:593.
-
- Sadler P.J., Muncie C., Shipman M.A. In: Biological Inorganic Chemistry: Structure & Reactivity. Bertini I., Gray H.B., Stiefel E.I., Valentine J.S., editors. University Science Books; Mill Valley, CA: 2007. p. 95.
-
- Park M., Li Q., Shcheynikov N., Muallem S., Zeng W. Cell Cycle. 2005;4:24. - PubMed
-
- Camilleri C., Markich S.J., Noller B.N., Turley C.J., Parker G., van Dam R.A. Chemosphere. 2002;50:355. - PubMed
-
- Levina A., Lay P.A. Coord. Chem. Rev. 2005;249:281.
Publication types
LinkOut - more resources
Full Text Sources
