Molecular Pathogenesis of Middle East Respiratory Syndrome (MERS) Coronavirus
- PMID: 32226718
- PMCID: PMC7100557
- DOI: 10.1007/s40588-019-00122-7
Molecular Pathogenesis of Middle East Respiratory Syndrome (MERS) Coronavirus
Abstract
Purpose of review: Middle East respiratory syndrome coronavirus (MERS-CoV) emerged in 2012 and is listed in the World Health Organization's blueprint of priority diseases that need immediate research. Camels are reservoirs of this virus, and the virus spills over into humans through direct contact with camels. Human-to-human transmission and travel-associated cases have been identified as well. Limited studies have characterized the molecular pathogenesis of MERS-CoV. Most studies have used ectopic expression of viral proteins to characterize MERS-CoV and its ability to modulate antiviral responses in human cells. Studies with live virus are limited, largely due to the requirement of high containment laboratories. In this review, we have summarized current studies on MERS-CoV molecular pathogenesis and have mentioned some recent strategies that are being developed to control MERS-CoV infection.
Recent findings: Multiple antiviral molecules with the potential to inhibit MERS-CoV infection by disrupting virus-receptor interactions are being developed and tested. Although human vaccine candidates are still being developed, a candidate camel vaccine is being tested for efficacy. Combination of supportive treatment with interferon and antivirals is also being explored.
Summary: New antiviral molecules that inhibit MERS-CoV and host cell receptor interaction may become available in the future. Additional studies are required to identify and characterize the pathogenesis of MERS-CoV EMC/2012 and other circulating strains. An effective MERS-CoV vaccine, for humans and/or camels, along with an efficient combination antiviral therapy may help us prevent future MERS cases.
Keywords: Coronavirus; MERS; Pathogenesis; Therapeutics.
© Springer Nature Switzerland AG 2019.
Conflict of interest statement
Conflict of InterestArinjay Banerjee and Karen Mossman each report that their lab is studying MERS-CoV and associated innate immune signaling in bat cells. Kaushal Baid declares no potential conflicts of interest.
Figures
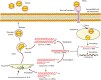

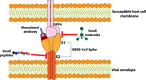
References
-
- CDC. Severe acute respiratory syndrome. 2004. https://www.cdc.gov/sars/about/fs-sars.html. Accessed February 20 2019.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
