Optimization of Synovial Fluid Collection and Processing for NMR Metabolomics and LC-MS/MS Proteomics
- PMID: 32227958
- PMCID: PMC7341532
- DOI: 10.1021/acs.jproteome.0c00035
Optimization of Synovial Fluid Collection and Processing for NMR Metabolomics and LC-MS/MS Proteomics
Abstract
Synovial fluid (SF) is of great interest for the investigation of orthopedic pathologies, as it is in close proximity to various tissues that are primarily altered during these disease processes and can be collected using minimally invasive protocols. Multi-"omic" approaches are commonplace, although little consideration is often given for multiple analysis techniques at sample collection. Nuclear magnetic resonance (NMR) metabolomics and liquid chromatography tandem mass spectrometry (LC-MS/MS) proteomics are two complementary techniques particularly suited to the study of SF. However, currently there are no agreed upon standard protocols that are published for SF collection and processing for use with NMR metabolomic analysis. Furthermore, the large protein concentration dynamic range present within SF can mask the detection of lower abundance proteins in proteomics. While combinational ligand libraries (ProteoMiner columns) have been developed to reduce this dynamic range, their reproducibility when used in conjunction with SF, or on-bead protein digestion protocols, has yet to be investigated. Here we employ optimized protocols for the collection, processing, and storage of SF for NMR metabolite analysis and LC-MS/MS proteome analysis, including a Lys-C endopeptidase digestion step prior to tryptic digestion, which increased the number of protein identifications and improved reproducibility for on-bead ProteoMiner digestion.
Keywords: Lys-C endopeptidase; mass spectrometry; metabolomics; nuclear magnetic resonance; proteomics; synovial fluid.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
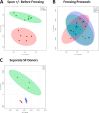


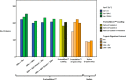
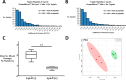
References
-
- Mateos J.; Lourido L.; Fernandez-Puente P.; Calamia V.; Fernandez-Lopez C.; Oreiro N.; Ruiz-Romero C.; Blanco F. J. Differential Protein Profiling of Synovial Fluid from Rheumatoid Arthritis and Osteoarthritis Patients Using LC-MALDI TOF/TOF. J. Proteomics 2012, 75 (10), 2869–2878. 10.1016/j.jprot.2011.12.042. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

