An integrated prognosis model of pharmacogenomic gene signature and clinical information for diffuse large B-cell lymphoma patients following CHOP-like chemotherapy
- PMID: 32228625
- PMCID: PMC7106727
- DOI: 10.1186/s12967-020-02311-1
An integrated prognosis model of pharmacogenomic gene signature and clinical information for diffuse large B-cell lymphoma patients following CHOP-like chemotherapy
Abstract
Background: As the most common form of lymphoma, diffuse large B-cell lymphoma (DLBCL) is a clinical highly heterogeneous disease with variability in therapeutic outcomes and biological features. It is a challenge to identify of clinically meaningful tools for outcome prediction. In this study, we developed a prognosis model fused clinical characteristics with drug resistance pharmacogenomic signature to identify DLBCL prognostic subgroups for CHOP-based treatment.
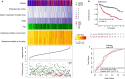
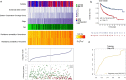
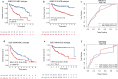
Methods: The expression microarray data and clinical characteristics of 791 DLBCL patients from two Gene Expression Omnibus (GEO) databases were used to establish and validate this model. By using univariate Cox regression, eight clinical or genetic signatures were analyzed. The elastic net-regulated Cox regression analysis was used to select the best prognosis related factors into the predictive model. To estimate the prognostic capability of the model, Kaplan-Meier curve and the area under receiver operating characteristic (ROC) curve (AUC) were performed.
Results: A predictive model comprising 4 clinical factors and 2 pharmacogenomic gene signatures was established after 1000 times cross validation in the training dataset. The AUC of the comprehensive risk model was 0.78, whereas AUC value was lower for the clinical only model (0.68) or the gene only model (0.67). Compared with low-risk patients, the overall survival (OS) of DLBCL patients with high-risk scores was significantly decreased (HR = 4.55, 95% CI 3.14-6.59, log-rank p value = 1.06 × 10-15). The signature also enables to predict prognosis within different molecular subtypes of DLBCL. The reliability of the integrated model was confirmed by independent validation dataset (HR = 3.47, 95% CI 2.42-4.97, log rank p value = 1.53 × 10-11).
Conclusions: This integrated model has a better predictive capability to ascertain the prognosis of DLBCL patients prior to CHOP-like treatment, which may improve the clinical management of DLBCL patients and provide theoretical basis for individualized treatment.
Keywords: CHOP-like chemotherapy; Diffuse large B-cell lymphoma; Pharmacogenomic signature; Survival.
Conflict of interest statement
None of the authors has any conflict of interest regarding this study.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

