Identification of genes required for the fitness of Streptococcus equi subsp. equi in whole equine blood and hydrogen peroxide
- PMID: 32228801
- PMCID: PMC7276704
- DOI: 10.1099/mgen.0.000362
Identification of genes required for the fitness of Streptococcus equi subsp. equi in whole equine blood and hydrogen peroxide
Abstract
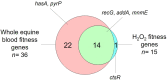
The availability of next-generation sequencing techniques provides an unprecedented opportunity for the assignment of gene function. Streptococcus equi subspecies equi is the causative agent of strangles in horses, one of the most prevalent and important diseases of equids worldwide. However, the live attenuated vaccines that are utilized to control this disease cause adverse reactions in some animals. Here, we employ transposon-directed insertion-site sequencing (TraDIS) to identify genes that are required for the fitness of S. equi in whole equine blood or in the presence of H2O2 to model selective pressures exerted by the equine immune response during infection. We report the fitness values of 1503 and 1471 genes, representing 94.5 and 92.5 % of non-essential genes in S. equi, following incubation in whole blood and in the presence of H2O2, respectively. Of these genes, 36 and 15 were identified as being important to the fitness of S. equi in whole blood or H2O2, respectively, with 14 genes being important in both conditions. Allelic replacement mutants were generated to validate the fitness results. Our data identify genes that are important for S. equi to resist aspects of the immune response in vitro, which can be exploited for the development of safer live attenuated vaccines to prevent strangles.
Keywords: Streptococcus equi; hydrogen peroxide; transposon-directed insertion-site sequencing; whole blood.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

