The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis
- PMID: 32231244
- PMCID: PMC7105485
- DOI: 10.1038/s41467-020-15383-w
The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis
Abstract
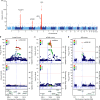
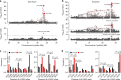
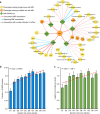
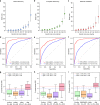
Membranous Nephropathy (MN) is a rare autoimmune cause of kidney failure. Here we report a genome-wide association study (GWAS) for primary MN in 3,782 cases and 9,038 controls of East Asian and European ancestries. We discover two previously unreported loci, NFKB1 (rs230540, OR = 1.25, P = 3.4 × 10-12) and IRF4 (rs9405192, OR = 1.29, P = 1.4 × 10-14), fine-map the PLA2R1 locus (rs17831251, OR = 2.25, P = 4.7 × 10-103) and report ancestry-specific effects of three classical HLA alleles: DRB1*1501 in East Asians (OR = 3.81, P = 2.0 × 10-49), DQA1*0501 in Europeans (OR = 2.88, P = 5.7 × 10-93), and DRB1*0301 in both ethnicities (OR = 3.50, P = 9.2 × 10-23 and OR = 3.39, P = 5.2 × 10-82, respectively). GWAS loci explain 32% of disease risk in East Asians and 25% in Europeans, and correctly re-classify 20-37% of the cases in validation cohorts that are antibody-negative by the serum anti-PLA2R ELISA diagnostic test. Our findings highlight an unusual genetic architecture of MN, with four loci and their interactions accounting for nearly one-third of the disease risk.
Trial registration: ClinicalTrials.gov NCT01180036.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Genetics of MN.Nat Rev Nephrol. 2020 Jun;16(6):316. doi: 10.1038/s41581-020-0282-9. Nat Rev Nephrol. 2020. PMID: 32265520 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

