Minimal phenotyping yields genome-wide association signals of low specificity for major depression
- PMID: 32231276
- PMCID: PMC7906795
- DOI: 10.1038/s41588-020-0594-5
Minimal phenotyping yields genome-wide association signals of low specificity for major depression
Abstract
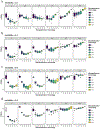
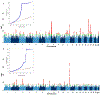
Minimal phenotyping refers to the reliance on the use of a small number of self-reported items for disease case identification, increasingly used in genome-wide association studies (GWAS). Here we report differences in genetic architecture between depression defined by minimal phenotyping and strictly defined major depressive disorder (MDD): the former has a lower genotype-derived heritability that cannot be explained by inclusion of milder cases and a higher proportion of the genome contributing to this shared genetic liability with other conditions than for strictly defined MDD. GWAS based on minimal phenotyping definitions preferentially identifies loci that are not specific to MDD, and, although it generates highly predictive polygenic risk scores, the predictive power can be explained entirely by large sample sizes rather than by specificity for MDD. Our results show that reliance on results from minimal phenotyping may bias views of the genetic architecture of MDD and impede the ability to identify pathways specific to MDD.
Figures











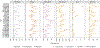

References
-
- Lu JT, Campeau PM & Lee BH Genotype–phenotype correlation: promiscuity in the era of next-generation sequencing. Obstet. Gynecol. Surv 69, 728–730 (2014). - PubMed

