Sestrins induce natural killer function in senescent-like CD8+ T cells
- PMID: 32231301
- PMCID: PMC10249464
- DOI: 10.1038/s41590-020-0643-3
Sestrins induce natural killer function in senescent-like CD8+ T cells
Abstract
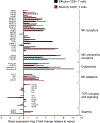
Aging is associated with remodeling of the immune system to enable the maintenance of life-long immunity. In the CD8+ T cell compartment, aging results in the expansion of highly differentiated cells that exhibit characteristics of cellular senescence. Here we found that CD27-CD28-CD8+ T cells lost the signaling activity of the T cell antigen receptor (TCR) and expressed a protein complex containing the agonistic natural killer (NK) receptor NKG2D and the NK adaptor molecule DAP12, which promoted cytotoxicity against cells that expressed NKG2D ligands. Immunoprecipitation and imaging cytometry indicated that the NKG2D-DAP12 complex was associated with sestrin 2. The genetic inhibition of sestrin 2 resulted in decreased expression of NKG2D and DAP12 and restored TCR signaling in senescent-like CD27-CD28-CD8+ T cells. Therefore, during aging, sestrins induce the reprogramming of non-proliferative senescent-like CD27-CD28-CD8+ T cells to acquire a broad-spectrum, innate-like killing activity.
Conflict of interest statement
Figures

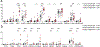


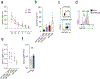




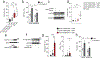


Comment in
-
Senescence blurs the line between innate and adaptive immune cells.Immunol Cell Biol. 2020 Jul;98(6):431-433. doi: 10.1111/imcb.12341. Epub 2020 May 13. Immunol Cell Biol. 2020. PMID: 32406096
References
-
- Akbar AN, Beverley PC & Salmon M Will telomere erosion lead to a loss of T-cell memory? Nature Reviews Immunology 4, nri1440 (2004). - PubMed
-
- Gray D A role for antigen in the maintenance of immunological memory. Nat Rev Immunol 2, nri706 (2002). - PubMed
-
- Mitri D et al. Reversible Senescence in Human CD4+CD45RA+CD27− Memory T Cells. The Journal of Immunology 187, 2093–2100 (2011). - PubMed
Methods references
Publication types
MeSH terms
Substances
Grants and funding
- R21 AG063528/AG/NIA NIH HHS/United States
- AZR00630/WT_/Wellcome Trust/United Kingdom
- MR/P00184X/1/MRC_/Medical Research Council/United Kingdom
- 26603/CRUK_/Cancer Research UK/United Kingdom
- R35 GM124922/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- MR/T015853/1/MRC_/Medical Research Council/United Kingdom
- R33 AG061456/AG/NIA NIH HHS/United States
- R01 AG048023/AG/NIA NIH HHS/United States
- R01 AI142086/AI/NIAID NIH HHS/United States
- R01 AG051647/AG/NIA NIH HHS/United States
- R01 DK111465/DK/NIDDK NIH HHS/United States
- R01 DK102850/DK/NIDDK NIH HHS/United States
- BB/L005336/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
- P01 AG021600/AG/NIA NIH HHS/United States
- R01 AG052608/AG/NIA NIH HHS/United States
- R56 AG060746/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

