A Systematic Bioinformatics Workflow With Meta-Analytics Identified Potential Pathogenic Factors of Alzheimer's Disease
- PMID: 32231518
- PMCID: PMC7083177
- DOI: 10.3389/fnins.2020.00209
A Systematic Bioinformatics Workflow With Meta-Analytics Identified Potential Pathogenic Factors of Alzheimer's Disease
Abstract
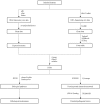
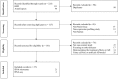
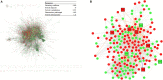
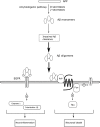
Potential pathogenic factors, other than well-known APP, APOE4, and PSEN, can be further identified from transcriptomics studies of differentially expressed genes (DEGs) that are specific for Alzheimer's disease (AD), but findings are often inconsistent or even contradictory. Evidence corroboration by combining meta-analysis and bioinformatics methods may help to resolve existing inconsistencies and contradictions. This study aimed to demonstrate a systematic workflow for evidence synthesis of transcriptomic studies using both meta-analysis and bioinformatics methods to identify potential pathogenic factors. Transcriptomic data were assessed from GEO and ArrayExpress after systematic searches. The DEGs and their dysregulation states from both DNA microarray and RNA sequencing datasets were analyzed and corroborated by meta-analysis. Statistically significant DEGs were used for enrichment analysis based on KEGG and protein-protein interaction network (PPIN) analysis based on STRING. AD-specific modules were further determined by the DIAMOnD algorithm, which identifies significant connectivity patterns between specific disease-associated proteins and non-specific proteins. Within AD-specific modules, the nodes of highest degrees (>95th percentile) were considered as potential pathogenic factors. After systematic searches of 225 datasets, extensive meta-analyses among 25 datasets (21 DNA microarray datasets and 4 RNA sequencing datasets) identified 9,298 DEGs. The dysregulated genes and pathways in AD were associated with impaired amyloid-β (Aβ) clearance. From the AD-specific module, Fyn, and EGFR were the most statistically significant and biologically relevant. This meta-analytical study suggested that the reduced Aβ clearance in AD pathogenesis was associated with the genes encoding Fyn and EGFR, which were key receptors in Aβ downstream signaling.
Keywords: Alzheimer’s disease; RNA sequence analysis; bioinformatics; meta-analysis; microarray analysis.
Copyright © 2020 Yuen, Zhu and Leung.
Figures




References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

