Identification of a General Odorant Receptor for Repellents in the Asian Corn Borer Ostrinia furnacalis
- PMID: 32231586
- PMCID: PMC7083148
- DOI: 10.3389/fphys.2020.00176
Identification of a General Odorant Receptor for Repellents in the Asian Corn Borer Ostrinia furnacalis
Abstract
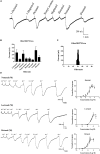
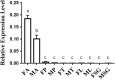
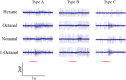
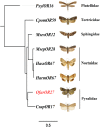
Attractants and repellents are considered to be an environment-friendly approach for pest management. Odorant receptors (ORs), which are located on the dendritic membranes of olfactory sensory neurons in insects, are essential genes for recognizing attractants and repellents. In the Asian corn borer, Ostrinia furnacalis, ORs that respond to sex pheromones have been characterized, but general ORs for plant odorants, especially for repellents, have not been identified. Nonanal is a plant volatile of maize that could result in avoidance of the oviposition process for female adults in O. furnacalis. In this study, we identified a female-biased OR that responds to nonanal using a Xenopus oocyte expression system. In addition, we found that OfurOR27 was also sensitive to two other compounds, octanal and 1-octanol. Behavioral analysis showed that octanal and 1-octanol also caused female avoidance of oviposition. Our results indicated that OfurOR27 is an OR that is sensitive to repellents. Moreover, the two newly identified repellents may help to develop a chemical ecology approach for pest control in O. furnacalis.
Keywords: heterologous expression system; host plant volatile; nonanal; odorant receptor; repellent.
Copyright © 2020 Yu, Yang, Chang, Zhang and Wang.
Figures


References
LinkOut - more resources
Full Text Sources

