Natural-Killer-Derived Extracellular Vesicles: Immune Sensors and Interactors
- PMID: 32231660
- PMCID: PMC7082405
- DOI: 10.3389/fimmu.2020.00262
Natural-Killer-Derived Extracellular Vesicles: Immune Sensors and Interactors
Abstract
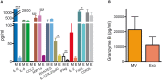
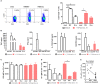
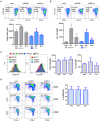
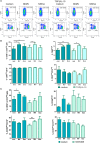
Natural killer (NK) cells contribute to immunosurveillance and first-line defense in the control of tumor growth and metastasis diffusion. NK-cell-derived extracellular vesicles (NKEVs) are constitutively secreted and biologically active. They reflect the protein and genetic repertoire of originating cells, and exert antitumor activity in vitro and in vivo. Cancer can compromise NK cell functions, a status potentially reflected by their extracellular vesicles. Hence, NKEVs could, on the one hand, contribute to improve cancer therapy by interacting with tumor and/or immune cells and on the other hand, sense the actual NK cell status in cancer patients. Here, we investigated the composition of healthy donors' NKEVs, including NK microvesicles and exosomes, and their interaction with uncompromised cells of the immune system. To sense the systemic NK cell status in cancer patients, we developed an immune enzymatic test (NKExoELISA) that measures plasma NK-cell-derived exosomes, captured as tsg101+CD56+ nanovesicles. NKEV mass spectrometry and cytokine analysis showed the expression of NK cell markers, i.e., NKG2D and CD94, perforin, granzymes, CD40L, and other molecules involved in cytotoxicity, homing, cell adhesion, and immune activation, together with EV markers tsg101, CD81, CD63, and CD9 in both NK-derived exosomes and microvesicles. Data are available via Proteome Xchange with identifier PXD014894. Immunomodulation studies revealed that NKEVs displayed main stimulatory functions in peripheral blood mononuclear cells (PBMCs), inducing the expression of human leukocyte antigen DR isotype (HLA-DR) and costimulatory molecules on monocytes and CD25 expression on T cells, which was maintained in the presence of lipopolysaccharide (LPS) and interleukin (IL)-10/transforming growth factor beta (TGFβ), respectively. Furthermore, NKEVs increased the CD56+ NK cell fraction, suggesting that effects mediated by NKEVs might be potentially exploited in support of cancer therapy. The measurement of circulating NK exosomes in the plasma of melanoma patients and healthy donors evidenced lower levels of tsg101+CD56+ exosomes in patients with respect to donors. Likewise, we detected lower frequencies of NK cells in PBMCs of these patients. These data highlight the potential of NKExoELISA to sense alterations of the NK cell immune status.
Keywords: exosomes; extracellular vesicles; healthy donors; immunosurveillance; melanoma patients; microvesicles; natural killer cells.
Copyright © 2020 Federici, Shahaj, Cecchetti, Camerini, Casella, Iessi, Camisaschi, Paolino, Calvieri, Ferro, Cova, Squarcina, Bertuccini, Iosi, Huber and Lugini.
Figures
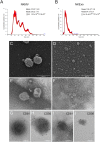

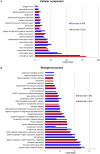
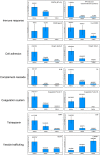
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

