Dental characters used in phylogenetic analyses of mammals show higher rates of evolution, but not reduced independence
- PMID: 32231876
- PMCID: PMC7100591
- DOI: 10.7717/peerj.8744
Dental characters used in phylogenetic analyses of mammals show higher rates of evolution, but not reduced independence
Abstract
Accurate reconstructions of phylogeny are essential for studying the evolution of a clade, and morphological characters are necessarily used for the reconstruction of the relationships of fossil organisms. However, variation in their evolutionary modes (for example rate variation and character non-independence) not accounted for in analyses may be leading to unreliable phylogenies. A recent study suggested that phylogenetic analyses of mammals may be suffering from a dominance of dental characters, which were shown to have lower phylogenetic signal than osteological characters and produced phylogenies less congruent with molecularly-derived benchmarks. Here we build on this previous work by testing five additional morphological partitions for phylogenetic signal and examining what aspects of dental and other character evolution may be affecting this, by fitting models of discrete character evolution to phylogenies inferred and time calibrated using molecular data. Results indicate that the phylogenetic signal of discrete characters correlate most strongly with rates of evolution, with increased rates driving increased homoplasy. In a dataset covering all Mammalia, dental characters have higher rates of evolution than other partitions. They do not, however, fit a model of independent character evolution any worse than other regions. Primates and marsupials show different patterns to other mammal clades, with dental characters evolving at slower rates and being more heavily integrated (less independent). While the dominance of dental characters in analyses of mammals could be leading to inaccurate phylogenies, the issue is not unique to dental characters and the results are not consistent across datasets. Molecular benchmarks (being entirely independent of the character data) provide a framework for examining each dataset individually to assess the evolution of the characters used.
Keywords: Evolutionary Rates; Homoplasy; Independence; Mammals; Phylogeny.
©2020 Brocklehurst and Benevento.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
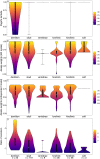

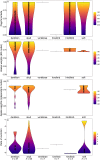
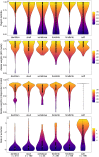
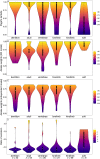
References
-
- Billet G, Bardin J. Serial homology and correlated characters in morphological phylogenetics: modelling the evolution of dental crests in placentals. Systematic Biology. 2018 (In Press) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

