Gastric Cancer Cell-Derived Exosomal microRNA-23a Promotes Angiogenesis by Targeting PTEN
- PMID: 32232005
- PMCID: PMC7082307
- DOI: 10.3389/fonc.2020.00326
Gastric Cancer Cell-Derived Exosomal microRNA-23a Promotes Angiogenesis by Targeting PTEN
Erratum in
-
Corrigendum: Gastric Cancer Cell-Derived Exosomal MicroRNA-23a Promotes Angiogenesis by Targeting PTEN.Front Oncol. 2022 Jan 20;11:797657. doi: 10.3389/fonc.2021.797657. eCollection 2021. Front Oncol. 2022. PMID: 35127509 Free PMC article.
Abstract
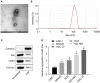
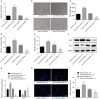
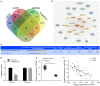
Hypoxia-exposed lung cancer-released exosomal microRNA-23a (miR-23a) has been shown to enhance angiogenesis as well as vascular permeability, contributing to the close correlation between exosomal miR-23a and tumorigenesis. The current study aimed to investigate whether gastric cancer (GC) cell-derived exosomal miR-23a could induce angiogenesis and to elucidate the potential mechanisms associated with the process. Differentially expressed miRNAs in GC were initially screened from the Gene Expression Omnibus database. Target genes were selected following miRNA-mRNA prediction and subsequently verified by dual luciferase reporter assay. RT-qPCR was conducted to detect miR-23a and PTEN expression in GC tissues, cells and exosomes. Human umbilical venous endothelial cells (HUVECs) were co-cultured with GC cell-derived exosomes to assess the angiogenesis mediated by exosomes in vitro. Additionally, PTEN was overexpressed in HUVECs to analyze the mechanism by which miR-23a regulates angiogenesis. miR-23a was highly expressed in GC tissues and cells and GC cell-derived exosomes. Angiogenesis was promoted by the co-culture of HUVECs and GC cells-derived exosomes, as evidenced by the increased expression of VEGF but decreased expression of TSP-1. PTEN was targeted by miR-23a and was lowly expressed in GC tissues. In a co-culture system, miR-23a carried by GC cells-derived exosomes promoted angiogenesis via the repression of PTEN. Collectively, GC cell-derived exosomal miR-23a could promote angiogenesis and provide blood supply for growth of GC cells. This study contributes to advancement of miRNA-targeted therapeutics.
Keywords: AKT pathway; PTEN; angiogenesis; exosome; gastric cancer; microRNA-23a.
Copyright © 2020 Du, Liang, Li, Zhao, Wang and Lin.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

