Rhinovirus Induces Basolateral Release of IL-17C in Highly Differentiated Airway Epithelial Cells
- PMID: 32232015
- PMCID: PMC7082745
- DOI: 10.3389/fcimb.2020.00103
Rhinovirus Induces Basolateral Release of IL-17C in Highly Differentiated Airway Epithelial Cells
Abstract
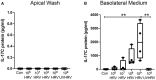
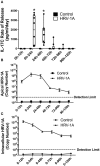
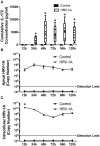
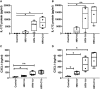
Human rhinovirus (HRV) is a major trigger of acute exacerbations of both asthma and chronic obstructive pulmonary disease. The airway epithelium is the primary site of HRV infection, and responds by releasing proinflammatory and antimicrobial cytokines. Epithelial cells release IL-17C in response to exposure to bacterial, viral, and fungal pathogens. We previously demonstrated a role for HRV in IL-17C production from undifferentiated epithelial cells, and showed that IL-17C could play a role in neutrophil recruitment. To extend these observations, highly differentiated human bronchial epithelial cells (HBE) were infected apically with HRV to assess the effect of dose, time, viral replication, and strain on the IL-17C response. Cellular lysates, and basolateral and apical secretions were analyzed for IL-17C and CXCL1 protein release following HRV or IL-17C stimulation. Upon HRV infection, IL-17C protein was exclusively released basolaterally in a dose-, time-, and viral replication-dependent manner. Several strains of rhinovirus were capable of inducing IL-17C release. Enriched columnar epithelial cell populations contained significantly higher viral titer, and expressed significantly more IL-17C mRNA than enriched basal cell populations. In addition, the kinetic profile of IL-17C release following HRV treatment closely mimics viral shedding kinetics, further implicating the role of rhinovirus replication in IL-17C production. Basolateral treatment of HBEs with IL-17C resulted in a dose-dependent increase in basolateral CXCL1 production. In summary, replicating rhinovirus drives basolateral IL-17C protein release from both apical and basal epithelial cells, which may then act in an autocrine/paracrine manner to promote basolateral CXCL1 protein release.
Keywords: IL-17C; air-liquid interface; airway epithelium; basolateral secretion; rhinovirus; well-differentiated.
Copyright © 2020 Jamieson, Wiehler, Michi and Proud.
Figures


Similar articles
-
Rhinovirus and Bacteria Synergistically Induce IL-17C Release from Human Airway Epithelial Cells To Promote Neutrophil Recruitment.J Immunol. 2019 Jan 1;202(1):160-170. doi: 10.4049/jimmunol.1800547. Epub 2018 Nov 30. J Immunol. 2019. PMID: 30504421
-
Interleukin-17A modulates human airway epithelial responses to human rhinovirus infection.Am J Physiol Lung Cell Mol Physiol. 2007 Aug;293(2):L505-15. doi: 10.1152/ajplung.00066.2007. Epub 2007 Jun 1. Am J Physiol Lung Cell Mol Physiol. 2007. PMID: 17545490
-
In vitro susceptibility to rhinovirus infection is greater for bronchial than for nasal airway epithelial cells in human subjects.J Allergy Clin Immunol. 2009 Jun;123(6):1384-90.e2. doi: 10.1016/j.jaci.2009.03.010. Epub 2009 May 9. J Allergy Clin Immunol. 2009. PMID: 19428098 Free PMC article.
-
Rhinovirus-Induced Modulation of Epithelial Phenotype: Role in Asthma.Viruses. 2020 Nov 19;12(11):1328. doi: 10.3390/v12111328. Viruses. 2020. PMID: 33227953 Free PMC article. Review.
-
Natural and experimental rhinovirus infections of the lower respiratory tract.Am J Respir Crit Care Med. 1995 Oct;152(4 Pt 2):S46-52. doi: 10.1164/ajrccm/152.4_Pt_2.S46. Am J Respir Crit Care Med. 1995. PMID: 7551413 Review.
Cited by
-
Mechanistic Insights into the Roles of the IL-17/IL-17R Families in Pancreatic Cancer.Int J Mol Sci. 2023 Aug 31;24(17):13539. doi: 10.3390/ijms241713539. Int J Mol Sci. 2023. PMID: 37686343 Free PMC article. Review.
-
Viral Infection and Airway Epithelial Immunity in Asthma.Int J Mol Sci. 2022 Aug 31;23(17):9914. doi: 10.3390/ijms23179914. Int J Mol Sci. 2022. PMID: 36077310 Free PMC article. Review.
-
Asymptomatic SARS-CoV-2 Infection Is Associated With Higher Levels of Serum IL-17C, Matrix Metalloproteinase 10 and Fibroblast Growth Factors Than Mild Symptomatic COVID-19.Front Immunol. 2022 Apr 5;13:821730. doi: 10.3389/fimmu.2022.821730. eCollection 2022. Front Immunol. 2022. PMID: 35479098 Free PMC article.
-
The IL-17 receptor IL-17RE mediates polyIC-induced exacerbation of experimental allergic asthma.Respir Res. 2020 Jul 8;21(1):176. doi: 10.1186/s12931-020-01434-9. Respir Res. 2020. PMID: 32641167 Free PMC article.
-
Actin cytoskeleton remodeling disrupts physical barriers to infection and presents entry receptors to respiratory syncytial virus.J Gen Virol. 2023 Nov;104(11):001923. doi: 10.1099/jgv.0.001923. J Gen Virol. 2023. PMID: 38015055 Free PMC article.
References
-
- Bochkov Y. A., Watters K., Ashraf S., Griggs T. F., Devries M. K., Jackson D. J., et al. . (2015). Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc. Natl. Acad. Sci. U.S.A. 112, 5485–5490. 10.1073/pnas.1421178112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

