Muscle atrophy-related myotube-derived exosomal microRNA in neuronal dysfunction: Targeting both coding and long noncoding RNAs
- PMID: 32233025
- PMCID: PMC7253071
- DOI: 10.1111/acel.13107
Muscle atrophy-related myotube-derived exosomal microRNA in neuronal dysfunction: Targeting both coding and long noncoding RNAs
Abstract
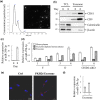
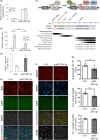
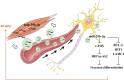
In mammals, microRNAs can be actively secreted from cells to blood. miR-29b-3p has been shown to play a pivotal role in muscle atrophy, but its role in intercellular communication is largely unknown. Here, we showed that miR-29b-3p was upregulated in normal and premature aging mouse muscle and plasma. miR-29b-3p was also upregulated in the blood of aging individuals, and circulating levels of miR-29b-3p were negatively correlated with relative appendicular skeletal muscle. Consistently, miR-29b-3p was observed in exosomes isolated from long-term differentiated atrophic C2C12 cells. When C2C12-derived miR-29b-3p-containing exosomes were uptaken by neuronal SH-SY5Y cells, increased miR-29b-3p levels in recipient cells were observed. Moreover, miR-29b-3p overexpression led to downregulation of neuronal-related genes and inhibition of neuronal differentiation. Interestingly, we identified HIF1α-AS2 as a novel c-FOS targeting lncRNA that is induced by miR-29b-3p through down-modulation of c-FOS and is required for miR-29b-3p-mediated neuronal differentiation inhibition. Our results suggest that atrophy-associated circulating miR-29b-3p may mediate distal communication between muscle cells and neurons.
Keywords: HIF-1α-AS2; aging; lncRNAs; miR-29b-3p; muscle atrophy.
© 2020 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures



References
-
- Arroyo, J. D. , Chevillet, J. R. , Kroh, E. M. , Ruf, I. K. , Pritchard, C. C. , Gibson, D. F. , … Tewari, M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences, 108, 5003–5008. 10.1073/pnas.1019055108 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

