Development of a prognostic index and screening of potential biomarkers based on immunogenomic landscape analysis of colorectal cancer
- PMID: 32235004
- PMCID: PMC7185108
- DOI: 10.18632/aging.102979
Development of a prognostic index and screening of potential biomarkers based on immunogenomic landscape analysis of colorectal cancer
Abstract
Background: Colorectal cancer (CRC) accounts for the highest fatality rate among all malignant tumors. Immunotherapy has shown great promise in management of many malignant tumors, necessitating the need to explore its role in CRC.
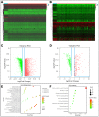
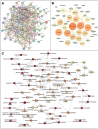
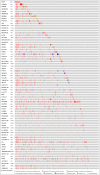
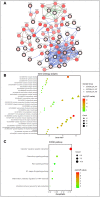
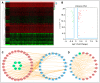
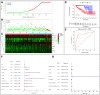
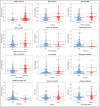
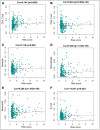
Results: Our analysis revealed a total of 71 differentially expressed IRGs, that were associated with prognosis of CRC patients. Ten IRGs (FABP4, IGKV1-33, IGKV2D-40, IGLV6-57, NGF, RETNLB, UCN, VIP, NGFR, and OXTR) showed high prognostic performance in predicting CRC outcomes, and were further associated with tumor burden, metastasis, tumor TNM stage, gender, age, and pathological stage. Interestingly, the IRG-based prognostic index (IRGPI) reflected infiltration of multiple immune cell types.
Conclusions: This model provides an effective approach for stratification and characterization of patients using IRG-based immunolabeling tools to monitor prognosis of CRC.
Methods: We performed a comprehensive analysis of expression profiles for immune-related genes (IRGs) and overall survival time in 437 CRC patients from the TCGA database. We employed computational algorithms and Cox regression analysis to estimate the relationship between differentially expressed IRGs and survival rates in CRC patients. Furthermore, we investigated the mechanisms of action of the IRGs involved in CRC, and established a novel prognostic index based on multivariate Cox models.
Keywords: colorectal cancer; immunogenomic landscape; personalized medicine; prognostic index; the cancer genome atlas.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

