Hyperspectral Imaging for the Detection of Glioblastoma Tumor Cells in H&E Slides Using Convolutional Neural Networks
- PMID: 32235483
- PMCID: PMC7181269
- DOI: 10.3390/s20071911
Hyperspectral Imaging for the Detection of Glioblastoma Tumor Cells in H&E Slides Using Convolutional Neural Networks
Abstract
Hyperspectral imaging (HSI) technology has demonstrated potential to provide useful information about the chemical composition of tissue and its morphological features in a single image modality. Deep learning (DL) techniques have demonstrated the ability of automatic feature extraction from data for a successful classification. In this study, we exploit HSI and DL for the automatic differentiation of glioblastoma (GB) and non-tumor tissue on hematoxylin and eosin (H&E) stained histological slides of human brain tissue. GB detection is a challenging application, showing high heterogeneity in the cellular morphology across different patients. We employed an HSI microscope, with a spectral range from 400 to 1000 nm, to collect 517 HS cubes from 13 GB patients using 20× magnification. Using a convolutional neural network (CNN), we were able to automatically detect GB within the pathological slides, achieving average sensitivity and specificity values of 88% and 77%, respectively, representing an improvement of 7% and 8% respectively, as compared to the results obtained using RGB (red, green, and blue) images. This study demonstrates that the combination of hyperspectral microscopic imaging and deep learning is a promising tool for future computational pathologies.
Keywords: convolutional neural networks; glioblastoma; hyperspectral imaging; medical optics and biotechnology; optical pathology; tissue characterization; tissue diagnostics.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

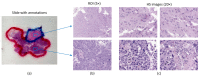



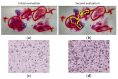
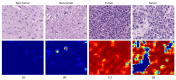

References
MeSH terms
Grants and funding
- RP190588/Cancer Prevention and Research Institute of Texas
- TEC2017-86722-C4-4-R/Ministerio de Economía, Industria y Competitividad, Gobierno de España
- ProID2017010164/Agencia Canaria de Investigación, Innovación y Sociedad de la Información
- TESIS2018010140/Agencia Canaria de Investigación, Innovación y Sociedad de la Información
LinkOut - more resources
Full Text Sources

