Molecular T-Cell Repertoire Analysis as Source of Prognostic and Predictive Biomarkers for Checkpoint Blockade Immunotherapy
- PMID: 32235561
- PMCID: PMC7177412
- DOI: 10.3390/ijms21072378
Molecular T-Cell Repertoire Analysis as Source of Prognostic and Predictive Biomarkers for Checkpoint Blockade Immunotherapy
Abstract
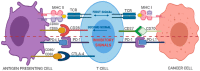
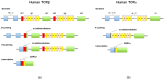
The T cells are key players of the response to checkpoint blockade immunotherapy (CBI) and monitoring the strength and specificity of antitumor T-cell reactivity remains a crucial but elusive component of precision immunotherapy. The entire assembly of T-cell receptor (TCR) sequences accounts for antigen specificity and strength of the T-cell immune response. The TCR repertoire hence represents a "footprint" of the conditions faced by T cells that dynamically evolves according to the challenges that arise for the immune system, such as tumor neo-antigenic load. Hence, TCR repertoire analysis is becoming increasingly important to comprehensively understand the nature of a successful antitumor T-cell response, and to improve the success and safety of current CBI.
Keywords: T-cell repertoire; TCR clonotype; checkpoint blockade immunotherapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Sosman J.A., Atkins M.B., Leming P.D., et al. Five-year survival and correlates among patients with advanced melanoma, renal cell carcinoma, or non-small cell lung cancer treated with nivolumab. [(accessed on 25 July 2019)];Jama Oncol. 2019 Available online: https://jamanetwork.com/journals/jamaoncology/fullarticle/2738775. - PMC - PubMed
-
- Masucci G.V., Cesano A., Hawtin R., Janetzki S., Zhang J., Kirsch I., Dobbin K.K., Alvarez J., Robbins P.B., Selvan S.R., et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume i-pre-analytical and analytical validation. J. Immunother. Cancer. 2016;4:76. doi: 10.1186/s40425-016-0178-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

