XPA: DNA Repair Protein of Significant Clinical Importance
- PMID: 32235701
- PMCID: PMC7139726
- DOI: 10.3390/ijms21062182
XPA: DNA Repair Protein of Significant Clinical Importance
Abstract
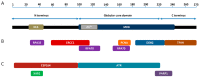
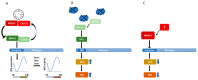
The nucleotide excision repair (NER) pathway is activated in response to a broad spectrum of DNA lesions, including bulky lesions induced by platinum-based chemotherapeutic agents. Expression levels of NER factors and resistance to chemotherapy has been examined with some suggestion that NER plays a role in tumour resistance; however, there is a great degree of variability in these studies. Nevertheless, recent clinical studies have suggested Xeroderma Pigmentosum group A (XPA) protein, a key regulator of the NER pathway that is essential for the repair of DNA damage induced by platinum-based chemotherapeutics, as a potential prognostic and predictive biomarker for response to treatment. XPA functions in damage verification step in NER, as well as a molecular scaffold to assemble other NER core factors around the DNA damage site, mediated by protein-protein interactions. In this review, we focus on the interacting partners and mechanisms of regulation of the XPA protein. We summarize clinical oncology data related to this DNA repair factor, particularly its relationship with treatment outcome, and examine the potential of XPA as a target for small molecule inhibitors.
Keywords: XPA protein; biomarker; cancer; nucleotide excision repair; prognostic and predictive value; single nucleotide polymorphism; small molecule inhibitors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

