Reciprocal regulation between alternative splicing and the DNA damage response
- PMID: 32236390
- PMCID: PMC7197977
- DOI: 10.1590/1678-4685-GMB-2019-0111
Reciprocal regulation between alternative splicing and the DNA damage response
Abstract
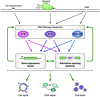
Splicing, the process that catalyzes intron removal and flanking exon ligation, can occur in different ways (alternative splicing) in immature RNAs transcribed from a single gene. In order to adapt to a particular context, cells modulate not only the quantity but also the quality (alternative isoforms) of their transcriptome. Since 95% of the human coding genome is subjected to alternative splicing regulation, it is expected that many cellular pathways are modulated by alternative splicing, as is the case for the DNA damage response. Moreover, recent evidence demonstrates that upon a genotoxic insult, classical DNA damage response kinases such as ATM, ATR and DNA-PK orchestrate the gene expression response therefore modulating alternative splicing which, in a reciprocal way, shapes the response to a damaging agent.
Conflict of interest statement
Figures
References
-
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. - PubMed
-
- Awasthi P, Foiani M, Kumar A. ATM and ATR signaling at a glance. J Cell Sci. 2015;128:4255–4262. - PubMed
-
- Baldin V, Cans C, Knibiehler M, Ducommun B. Phosphorylation of human CDC25B phosphatase by CDK1-cyclin A triggers its proteasome-dependent degradation. J Biol Chem. 1997;272:32731–3274. - PubMed
-
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

