EMT related circular RNA expression profiles identify circSCYL2 as a novel molecule in breast tumor metastasis
- PMID: 32236616
- PMCID: PMC7169655
- DOI: 10.3892/ijmm.2020.4550
EMT related circular RNA expression profiles identify circSCYL2 as a novel molecule in breast tumor metastasis
Abstract
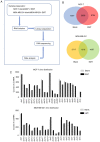
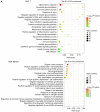
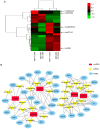
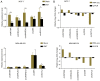
Substantial evidence indicates that circular RNAs (circRNAs) play vital roles in several diseases, especially in cancer development. However, the functions of circRNAs in breast cancer metastasis remain to be investigated. This study aimed to identify the key circRNAs involved in epithelial mesenchymal transition (EMT) of breast cancer and evaluated their molecular function and roles in pathways that may be associated with tumor metastasis. An EMT model was constructed by treating breast cancer cells MCF‑7 and MDA‑MB‑231 with transforming growth factor‑β1. High‑throughput RNA sequencing was used to identify the differentially expressed circRNAs in EMT and blank groups of two cells, and reverse transcription‑quantitative PCR was used to validate the expression of circSCYL2 in human breast cancer tissues and cells. The effects of circSCYL2 on breast cancer cells were explored by transfecting with plasmids and the biological roles were assessed using transwell assays. EMT groups of breast cancer cells exhibited the characteristics of mesenchymal cells. Furthermore, the present study found that 7 circRNAs were significantly upregulated in both the MCF‑7 EMT and MDA‑MB‑231 EMT groups, while 16 circRNAs were significantly downregulated. The current study identified that circSCYL2 was downregulated in breast cancer tissues and cell lines, and that circSCYL2 overexpression inhibited cell migration and invasion. This study provides expression profiles of circRNAs in EMT groups of breast cancer cells. circSCYL2, which is downregulated in breast cancer tissues and cells, may play an important role in breast cancer EMT progression.
Figures




References
-
- Rodgers RJ, Reid GD, Koch J, Deans R, Ledger WL, Friedlander M, Gilchrist RB, Walters KA, Abbott JA. The safety and efficacy of controlled ovarian hyperstimulation for fertility preservation in women with early breast cancer: A systematic review. Hum Reprod. 2017;32:1033–1045. doi: 10.1093/humrep/dex027. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

