In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel
- PMID: 32238078
- PMCID: PMC7171389
- DOI: 10.1080/07391102.2020.1751300
In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel
Abstract
Recent outbreak of Coronavirus disease (COVID-19) pandemic around the world is associated with 'severe acute respiratory syndrome' (SARS-CoV2) in humans. SARS-CoV2 is an enveloped virus and E proteins present in them are reported to form ion channels, which is mainly associated with pathogenesis. Thus, there is always a quest to inhibit these ion channels, which in turn may help in controlling diseases caused by SARS-CoV2 in humans. Considering this, in the present study, authors employed computational approaches for studying the structure as well as function of the human 'SARS-CoV2 E' protein as well as its interaction with various phytochemicals. Result obtained revealed that α-helix and loops present in this protein experience random movement under optimal condition, which in turn modulate ion channel activity; thereby aiding the pathogenesis caused via SARS-CoV2 in human and other vertebrates. However, after binding with Belachinal, Macaflavanone E, and Vibsanol B, the random motion of the human 'SARS-CoV2 E' protein gets reduced, this, in turn, inhibits the function of the 'SARS-CoV2 E' protein. It is pertinent to note that two amino acids, namely VAL25 and PHE26, play a key role while interacting with these three phytochemicals. As these three phytochemicals, namely, Belachinal, Macaflavanone E & Vibsanol B, have passed the ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) property as well as 'Lipinski's Rule of 5s', they may be utilized as drugs in controlling disease caused via SARS-COV2, after further investigation.Communicated by Ramaswamy H. Sarma.
Keywords: Coronaviruses; docking; modeling; molecular dynamics; severe acute respiratory syndrome.
Figures


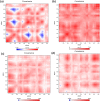

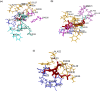
References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., & Lindahl E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. doi:10.1016/j.softx.2015.06.001 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
