Infant High-Grade Gliomas Comprise Multiple Subgroups Characterized by Novel Targetable Gene Fusions and Favorable Outcomes
- PMID: 32238360
- PMCID: PMC8313225
- DOI: 10.1158/2159-8290.CD-19-1030
Infant High-Grade Gliomas Comprise Multiple Subgroups Characterized by Novel Targetable Gene Fusions and Favorable Outcomes
Abstract
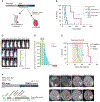
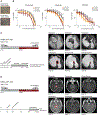
Infant high-grade gliomas appear clinically distinct from their counterparts in older children, indicating that histopathologic grading may not accurately reflect the biology of these tumors. We have collected 241 cases under 4 years of age, and carried out histologic review, methylation profiling, and custom panel, genome, or exome sequencing. After excluding tumors representing other established entities or subgroups, we identified 130 cases to be part of an "intrinsic" spectrum of disease specific to the infant population. These included those with targetable MAPK alterations, and a large proportion of remaining cases harboring gene fusions targeting ALK (n = 31), NTRK1/2/3 (n = 21), ROS1 (n = 9), and MET (n = 4) as their driving alterations, with evidence of efficacy of targeted agents in the clinic. These data strongly support the concept that infant gliomas require a change in diagnostic practice and management. SIGNIFICANCE: Infant high-grade gliomas in the cerebral hemispheres comprise novel subgroups, with a prevalence of ALK, NTRK1/2/3, ROS1, or MET gene fusions. Kinase fusion-positive tumors have better outcome and respond to targeted therapy clinically. Other subgroups have poor outcome, with fusion-negative cases possibly representing an epigenetically driven pluripotent stem cell phenotype.See related commentary by Szulzewsky and Cimino, p. 904.This article is highlighted in the In This Issue feature, p. 890.
©2020 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the other authors.
Figures





Comment in
-
Fusing the Genetic Landscape of Infantile High-Grade Gliomas.Cancer Discov. 2020 Jul;10(7):904-906. doi: 10.1158/2159-8290.CD-20-0495. Cancer Discov. 2020. PMID: 32611733
References
-
- Jones C, Perryman L, Hargrave D . Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol 2012;9: 400–13. - PubMed
-
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31. - PubMed
-
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425–37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

