Centrosome anchoring regulates progenitor properties and cortical formation
- PMID: 32238932
- PMCID: PMC7138347
- DOI: 10.1038/s41586-020-2139-6
Centrosome anchoring regulates progenitor properties and cortical formation
Abstract
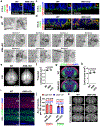
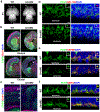
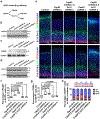
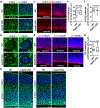
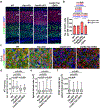
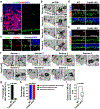
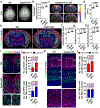
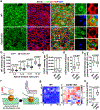
Radial glial progenitor cells (RGPs) are the major neural progenitor cells that generate neurons and glia in the developing mammalian cerebral cortex1-4. In RGPs, the centrosome is positioned away from the nucleus at the apical surface of the ventricular zone of the cerebral cortex5-8. However, the molecular basis and precise function of this distinctive subcellular organization of the centrosome are largely unknown. Here we show in mice that anchoring of the centrosome to the apical membrane controls the mechanical properties of cortical RGPs, and consequently their mitotic behaviour and the size and formation of the cortex. The mother centriole in RGPs develops distal appendages that anchor it to the apical membrane. Selective removal of centrosomal protein 83 (CEP83) eliminates these distal appendages and disrupts the anchorage of the centrosome to the apical membrane, resulting in the disorganization of microtubules and stretching and stiffening of the apical membrane. The elimination of CEP83 also activates the mechanically sensitive yes-associated protein (YAP) and promotes the excessive proliferation of RGPs, together with a subsequent overproduction of intermediate progenitor cells, which leads to the formation of an enlarged cortex with abnormal folding. Simultaneous elimination of YAP suppresses the cortical enlargement and folding that is induced by the removal of CEP83. Together, these results indicate a previously unknown role of the centrosome in regulating the mechanical features of neural progenitor cells and the size and configuration of the mammalian cerebral cortex.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

