Nanoscale imaging of clinical specimens using conventional and rapid-expansion pathology
- PMID: 32238952
- PMCID: PMC7490752
- DOI: 10.1038/s41596-020-0300-1
Nanoscale imaging of clinical specimens using conventional and rapid-expansion pathology
Abstract
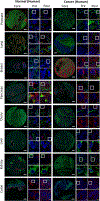
In pathology, microscopy is an important tool for the analysis of human tissues, both for the scientific study of disease states and for diagnosis. However, the microscopes commonly used in pathology are limited in resolution by diffraction. Recently, we discovered that it was possible, through a chemical process, to isotropically expand preserved cells and tissues by 4-5× in linear dimension. We call this process expansion microscopy (ExM). ExM enables nanoscale resolution imaging on conventional microscopes. Here we describe protocols for the simple and effective physical expansion of a variety of human tissues and clinical specimens, including paraffin-embedded, fresh frozen and chemically stained human tissues. These protocols require only inexpensive, commercially available reagents and hardware commonly found in a routine pathology laboratory. Our protocols are written for researchers and pathologists experienced in conventional fluorescence microscopy. The conventional protocol, expansion pathology, can be completed in ~1 d with immunostained tissue sections and 2 d with unstained specimens. We also include a new, fast variant, rapid expansion pathology, that can be performed on <5-µm-thick tissue sections, taking <4 h with immunostained tissue sections and <8 h with unstained specimens.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors have filed and obtained patent protection on a subset of the technologies here described (US provisional application no. 62/299,754, 62/463,265 and 62/463,251). E.S.B. helped cofound a company to help disseminate expansion microscopy to the community. OB is the co-Founder and CEO of QPathology LLC, Boston, MA.
Figures

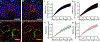





References
-
- Hell SW Far-field optical nanoscopy. in 2010 23rd Annual Meeting of the IEEE Photonics Society, PHOTINICS 2010 3–4 (2010). doi:10.1109/PHOTONICS.2010.5698725 - DOI
-
- Betzig E Proposed method for molecular optical imaging. Opt. Lett. 20, 237–9 (1995). - PubMed
-
- Betzig E et al. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 313, 1642–1645 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

