Application of Combined Genomic and Transfer Analyses to Identify Factors Mediating Regional Spread of Antibiotic-resistant Bacterial Lineages
- PMID: 32239131
- PMCID: PMC7745002
- DOI: 10.1093/cid/ciaa364
Application of Combined Genomic and Transfer Analyses to Identify Factors Mediating Regional Spread of Antibiotic-resistant Bacterial Lineages
Abstract
Background: Patients entering nursing facilities (NFs) are frequently colonized with antibiotic-resistant organisms (AROs). To understand the determinants of ARO colonization on NF admission, we applied whole-genome sequencing to track the spread of 4 ARO species across regional NFs and evaluated patient-level characteristics and transfer acute care hospitals (ACHs) as risk factors for colonization.
Methods: Patients from 6 NFs (n = 584) were surveyed for methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecalis/faecium (VREfc/VREfm), and ciprofloxacin-resistant Escherichia coli (CipREc) colonization. Genomic analysis was performed to quantify ARO spread between NFs and compared to patient-transfer networks. The association between admission colonization and patient-level variables and recent ACH exposures was examined.
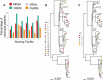
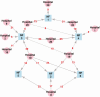
Results: The majority of ARO isolates belonged to major healthcare-associated lineages: MRSA (sequence type [ST] 5); VREfc (ST6); CipREc (ST131), and VREfm (clade A). While the genomic similarity of strains between NF pairs was positively associated with overlap in their feeder ACHs (P < .05 for MRSA, VREfc, and CipREc), limited phylogenetic clustering by either ACH or NF supported regional endemicity. Significant predictors for ARO colonization on NF admission included lower functional status and recent exposure to glycopeptides (adjusted odds ratio [aOR], > 2 for MRSA and VREfc/VREfm) or third-/fourth-generation cephalosporins (aOR, > 2 for MRSA and VREfm). Transfer from specific ACHs was an independent risk factor for only 1 ARO/ACH pair (VREfm/ACH19: aOR, 2.48).
Conclusions: In this region, healthcare-associated ARO lineages are endemic among connected NFs and ACHs, making patient characteristics more informative of NF admission colonization risk than exposure to specific ACHs.
Keywords: antibiotic-resistant organisms; genomic epidemiology; nursing facilities; surveillance; transmission.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures

References
-
- Safdar N, Maki DG. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, Enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann Intern Med 2002; 136:834–44. - PubMed
-
- Burke RE, Juarez-Colunga E, Levy C, Prochazka AV, Coleman EA, Ginde AA. Rise of post–acute care facilities as a discharge destination of US hospitalizations. JAMA Intern Med 2015; 175:295–6. - PubMed
-
- Jenq GY, Tinetti ME. Post–acute care: who belongs where? JAMA Intern Med 2015; 175:296–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

