Multiview learning for understanding functional multiomics
- PMID: 32240163
- PMCID: PMC7117667
- DOI: 10.1371/journal.pcbi.1007677
Multiview learning for understanding functional multiomics
Abstract
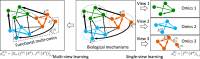
The molecular mechanisms and functions in complex biological systems currently remain elusive. Recent high-throughput techniques, such as next-generation sequencing, have generated a wide variety of multiomics datasets that enable the identification of biological functions and mechanisms via multiple facets. However, integrating these large-scale multiomics data and discovering functional insights are, nevertheless, challenging tasks. To address these challenges, machine learning has been broadly applied to analyze multiomics. This review introduces multiview learning-an emerging machine learning field-and envisions its potentially powerful applications to multiomics. In particular, multiview learning is more effective than previous integrative methods for learning data's heterogeneity and revealing cross-talk patterns. Although it has been applied to various contexts, such as computer vision and speech recognition, multiview learning has not yet been widely applied to biological data-specifically, multiomics data. Therefore, this paper firstly reviews recent multiview learning methods and unifies them in a framework called multiview empirical risk minimization (MV-ERM). We further discuss the potential applications of each method to multiomics, including genomics, transcriptomics, and epigenomics, in an aim to discover the functional and mechanistic interpretations across omics. Secondly, we explore possible applications to different biological systems, including human diseases (e.g., brain disorders and cancers), plants, and single-cell analysis, and discuss both the benefits and caveats of using multiview learning to discover the molecular mechanisms and functions of these systems.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- de Sa VR. Learning classification with unlabeled data. In: Advances in neural information processing systems. [Internet]. NIPS 1993. 1994 [cited 2020 Mar 17]. p. 112–119. Available from: https://papers.nips.cc/paper/831-learning-classification-with-unlabeled-...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

