Nucleotide sequence and analysis of pRC12 and pRC18, two theta-replicating plasmids harbored by Lactobacillus curvatus CRL 705
- PMID: 32240216
- PMCID: PMC7117683
- DOI: 10.1371/journal.pone.0230857
Nucleotide sequence and analysis of pRC12 and pRC18, two theta-replicating plasmids harbored by Lactobacillus curvatus CRL 705
Abstract
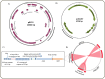
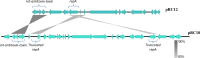
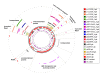
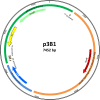
The nucleotide sequences of plasmids pRC12 (12,342 bp; GC 43.99%) and pRC18 (18,664 bp; GC 34.33%), harbored by the bacteriocin-producer Lactobacillus curvatus CRL 705, were determined and analyzed. Plasmids pRC12 and pRC18 share a region with high DNA identity (> 83% identity between RepA, a Type II toxin-antitoxin system and a tyrosine integrase genes) and are stably maintained in their natural host L. curvatus CRL 705. Both plasmids are low copy number and belong to the theta-type replicating group. While pRC12 is a pUCL287-like plasmid that possesses iterons and the repA and repB genes for replication, pRC18 harbors a 168 amino acid replication protein affiliated to RepB, which was named RepB'. Plasmid pRC18 also possesses a pUCL287-like repA gene but it was disrupted by an 11 kb insertion element that contains RepB', several transposases/IS elements, and the lactocin Lac705 operon. An Escherichia coli / Lactobacillus shuttle vector, named plasmid p3B1, carrying the pRC18 replicon (i.e. repB' and replication origin), a chloramphenicol resistance gene and a pBluescript backbone, was constructed and used to define the host range of RepB'. Chloramphenicol-resistant transformants were obtained after electroporation of Lactobacillus plantarum CRL 691, Lactobacillus sakei 23K and a plasmid-cured derivative of L. curvatus CRL 705, but not of L. curvatus DSM 20019 or Lactococcus lactis NZ9000. Depending on the host, transformation efficiency ranged from 102 to 107 per μg of DNA; in the new hosts, the plasmid was relatively stable as 29-53% of recombinants kept it after cell growth for 100 generations in the absence of selective pressure. Plasmid p3B1 could therefore be used for cloning and functional studies in several Lactobacillus species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- da Costa RJ, Voloski FLS, Mondadori RG, Duval EH, Fiorentini AM. Preservation of meat products with bacteriocins produced by lactic acid bacteria isolated from meat. J Food Qual. 2019, 1–12. 10.1155/2019/4726510 - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

