Genetic deletion of microRNA biogenesis in muscle cells reveals a hierarchical non-clustered network that controls focal adhesion signaling during muscle regeneration
- PMID: 32240622
- PMCID: PMC7139120
- DOI: 10.1016/j.molmet.2020.02.010
Genetic deletion of microRNA biogenesis in muscle cells reveals a hierarchical non-clustered network that controls focal adhesion signaling during muscle regeneration
Abstract
Objective: Decreased muscle mass is a major contributor to age-related morbidity, and strategies to improve muscle regeneration during ageing are urgently needed. Our aim was to identify the subset of relevant microRNAs (miRNAs) that partake in critical aspects of muscle cell differentiation, irrespective of computational predictions, genomic clustering or differential expression of the miRNAs.
Methods: miRNA biogenesis was deleted in primary myoblasts using a tamoxifen-inducible CreLox system and combined with an add-back miRNA library screen. RNA-seq experiments, cellular signalling events, and glycogen synthesis, along with miRNA inhibitors, were performed in human primary myoblasts. Muscle regeneration in young and aged mice was assessed using the cardiotoxin (CTX) model.
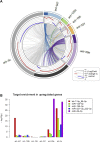
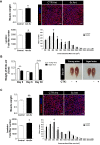
Results: We identified a hierarchical non-clustered miRNA network consisting of highly (miR-29a), moderately (let-7) and mildly active (miR-125b, miR-199a, miR-221) miRNAs that cooperate by directly targeting members of the focal adhesion complex. Through RNA-seq experiments comparing single versus combinatorial inhibition of the miRNAs, we uncovered a fundamental feature of this network, that miRNA activity inversely correlates to miRNA cooperativity. During myoblast differentiation, combinatorial inhibition of the five miRNAs increased activation of focal adhesion kinase (FAK), AKT, and p38 mitogen-activated protein kinase (MAPK), and improved myotube formation and insulin-dependent glycogen synthesis. Moreover, antagonizing the miRNA network in vivo following CTX-induced muscle regeneration enhanced muscle mass and myofiber formation in young and aged mice.
Conclusion: Our results provide novel insights into the dynamics of miRNA cooperativity and identify a miRNA network as therapeutic target for impaired focal adhesion signalling and muscle regeneration during ageing.
Keywords: Focal adhesion signalling; Glycogen synthesis; Primary human muscle cells; Skeletal muscle regeneration; microRNA network.
Copyright © 2020 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures







Similar articles
-
Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis.J Cell Sci. 2013 Jun 15;126(Pt 12):2678-91. doi: 10.1242/jcs.119966. Epub 2013 Apr 19. J Cell Sci. 2013. PMID: 23606743 Free PMC article.
-
MicroRNA-29a in Adult Muscle Stem Cells Controls Skeletal Muscle Regeneration During Injury and Exercise Downstream of Fibroblast Growth Factor-2.Stem Cells. 2016 Mar;34(3):768-80. doi: 10.1002/stem.2281. Epub 2016 Jan 29. Stem Cells. 2016. PMID: 26731484
-
miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4.Genes Dev. 2015 Aug 1;29(15):1605-17. doi: 10.1101/gad.263574.115. Epub 2015 Jul 27. Genes Dev. 2015. PMID: 26215566 Free PMC article.
-
Progress on the regulation of muscle regeneration by microRNA.Yi Chuan. 2025 May;47(5):513-532. doi: 10.16288/j.yczz.24-252. Yi Chuan. 2025. PMID: 40287786 Review.
-
Non-Coding RNAs as Regulators of Myogenesis and Postexercise Muscle Regeneration.Int J Mol Sci. 2021 Oct 26;22(21):11568. doi: 10.3390/ijms222111568. Int J Mol Sci. 2021. PMID: 34768999 Free PMC article. Review.
Cited by
-
Possible Combinatorial Utilization of Phytochemicals and Extracellular Vesicles for Wound Healing and Regeneration.Int J Mol Sci. 2024 Sep 26;25(19):10353. doi: 10.3390/ijms251910353. Int J Mol Sci. 2024. PMID: 39408681 Free PMC article. Review.
-
Older postmenopausal women with lower lean mass have hypermethylated sites in the PI3K-Akt pathway.Front Physiol. 2023 Apr 10;14:1150821. doi: 10.3389/fphys.2023.1150821. eCollection 2023. Front Physiol. 2023. PMID: 37123284 Free PMC article.
-
MicroRNA profiling of different exercise interventions for alleviating skeletal muscle atrophy in naturally aging rats.J Cachexia Sarcopenia Muscle. 2023 Feb;14(1):356-368. doi: 10.1002/jcsm.13137. Epub 2022 Dec 1. J Cachexia Sarcopenia Muscle. 2023. PMID: 36457259 Free PMC article.
-
Skeletal Muscle Regeneration in Cardiotoxin-Induced Muscle Injury Models.Int J Mol Sci. 2022 Nov 2;23(21):13380. doi: 10.3390/ijms232113380. Int J Mol Sci. 2022. PMID: 36362166 Free PMC article. Review.
-
FGF-2-dependent signaling activated in aged human skeletal muscle promotes intramuscular adipogenesis.Proc Natl Acad Sci U S A. 2021 Sep 14;118(37):e2021013118. doi: 10.1073/pnas.2021013118. Proc Natl Acad Sci U S A. 2021. PMID: 34493647 Free PMC article.
References
-
- Myers J., Prakash M., Froelicher V., Do D., Partington S., Atwood J.E. Exercise capacity and mortality among men referred for exercise testing. New England Journal of Medicine. 2002;346(11):793–801. - PubMed
-
- Shulman G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. New England Journal of Medicine. 2014;371(23):2237–2238. - PubMed
-
- Rosenberg I.H. Sarcopenia: origins and clinical relevance. Journal of Nutrition. 1997;127(5 Suppl):990S–991S. - PubMed
-
- Baumgartner R.N., Koehler K.M., Gallagher D., Romero L., Heymsfield S.B., Ross R.R. Epidemiology of sarcopenia among the elderly in New Mexico. American Journal of Epidemiology. 1998;147(8):755–763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

