The C-terminal tails of the mitochondrial transcription factors Mtf1 and TFB2M are part of an autoinhibitory mechanism that regulates DNA binding
- PMID: 32241911
- PMCID: PMC7242694
- DOI: 10.1074/jbc.RA120.013338
The C-terminal tails of the mitochondrial transcription factors Mtf1 and TFB2M are part of an autoinhibitory mechanism that regulates DNA binding
Abstract
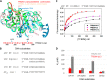
The structurally homologous Mtf1 and TFB2M proteins serve as transcription initiation factors of mitochondrial RNA polymerases in Saccharomyces cerevisiae and humans, respectively. These transcription factors directly interact with the nontemplate strand of the transcription bubble to drive promoter melting. Given the key roles of Mtf1 and TFB2M in promoter-specific transcription initiation, it can be expected that the DNA binding activity of the mitochondrial transcription factors is regulated to prevent DNA binding at inappropriate times. However, little information is available on how mitochondrial DNA transcription is regulated. While studying C-terminal (C-tail) deletion mutants of Mtf1 and TFB2M, we stumbled upon a finding that suggested that the flexible C-tail region of these factors autoregulates their DNA binding activity. Quantitative DNA binding studies with fluorescence anisotropy-based titrations revealed that Mtf1 with an intact C-tail has no affinity for DNA but deletion of the C-tail greatly increases Mtf1's DNA binding affinity. Similar observations were made with TFB2M, although autoinhibition by the C-tail of TFB2M was not as complete as in Mtf1. Analysis of available TFB2M structures disclosed that the C-tail engages in intramolecular interactions with the DNA binding groove in the free factor, which, we propose, inhibits its DNA binding activity. Further experiments showed that RNA polymerase relieves this autoinhibition by interacting with the C-tail and engaging it in complex formation. In conclusion, our biochemical and structural analyses reveal autoinhibitory and activation mechanisms of mitochondrial transcription factors that regulate their DNA binding activities and aid in specific assembly of transcription initiation complexes.
Keywords: Mtf1; RNA polymerase; TFB2M; autoinhibition; fluorescence anisotropy; mitochondria; mitochondrial RNA polymerase; mitochondrial transcription factors; protein–DNA interaction; transcriptional coactivator.
© 2020 Basu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures




References
-
- Jang S. H., and Jaehning J. A. (1991) The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J. Biol. Chem. 266, 22671–22677 - PubMed
-
- Basu U., Lee S. W., Deshpande A., Shen J., Sohn B. K., Cho H., Kim H., and Patel S. S. (2020) The C-terminal tail of the yeast mitochondrial transcription factor Mtf1 coordinates template strand alignment, DNA scrunching and timely transition into elongation. Nucleic Acids Res. 48, 2604–2620 10.1093/nar/gkaa040 - DOI - PMC - PubMed
-
- Litonin D., Sologub M., Shi Y., Savkina M., Anikin M., Falkenberg M., Gustafsson C. M., and Temiakov D. (2010) Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285, 18129–18133 10.1074/jbc.C110.128918 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

