Dual functions of Aire CARD multimerization in the transcriptional regulation of T cell tolerance
- PMID: 32242017
- PMCID: PMC7118133
- DOI: 10.1038/s41467-020-15448-w
Dual functions of Aire CARD multimerization in the transcriptional regulation of T cell tolerance
Abstract
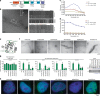
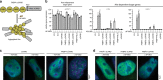
Aggregate-like biomolecular assemblies are emerging as new conformational states with functionality. Aire, a transcription factor essential for central T cell tolerance, forms large aggregate-like assemblies visualized as nuclear foci. Here we demonstrate that Aire utilizes its caspase activation recruitment domain (CARD) to form filamentous homo-multimers in vitro, and this assembly mediates foci formation and transcriptional activity. However, CARD-mediated multimerization also makes Aire susceptible to interaction with promyelocytic leukemia protein (PML) bodies, sites of many nuclear processes including protein quality control of nuclear aggregates. Several loss-of-function Aire mutants, including those causing autoimmune polyendocrine syndrome type-1, form foci with increased PML body association. Directing Aire to PML bodies impairs the transcriptional activity of Aire, while dispersing PML bodies with a viral antagonist restores this activity. Our study thus reveals a new regulatory role of PML bodies in Aire function, and highlights the interplay between nuclear aggregate-like assemblies and PML-mediated protein quality control.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

