Genetically similar temperate phages form coalitions with their shared host that lead to niche-specific fitness effects
- PMID: 32242083
- PMCID: PMC7305329
- DOI: 10.1038/s41396-020-0637-z
Genetically similar temperate phages form coalitions with their shared host that lead to niche-specific fitness effects
Abstract
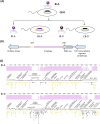
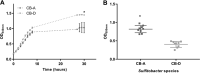
Temperate phages engage in long-term associations with their hosts that may lead to mutually beneficial interactions, of which the full extent is presently unknown. Here, we describe an environmentally relevant model system with a single host, a species of the Roseobacter clade of marine bacteria, and two genetically similar phages (ɸ-A and ɸ-D). Superinfection of a ɸ-D lysogenized strain (CB-D) with ɸ-A particles resulted in a lytic infection, prophage induction, and conversion of a subset of the host population, leading to isolation of a newly ɸ-A lysogenized strain (CB-A). Phenotypic differences, predicted to result from divergent lysogenic-lytic switch mechanisms, are evident between these lysogens, with CB-A displaying a higher incidence of spontaneous induction. Doubling times of CB-D and CB-A in liquid culture are 75 and 100 min, respectively. As cell cultures enter stationary phase, CB-A viable counts are half of CB-D. Consistent with prior evidence that cell lysis enhances biofilm formation, CB-A produces twice as much biofilm biomass as CB-D. As strains are susceptible to infection by the opposing phage type, co-culture competitions were performed to test fitness effects. When grown planktonically, CB-A outcompeted CB-D three to one. Yet, during biofilm growth, CB-D outcompeted CB-A three to one. These results suggest that genetically similar phages can have divergent influence on the competitiveness of their shared hosts in distinct environmental niches, possibly due to a complex form of phage-mediated allelopathy. These findings have implications for enhanced understanding of the eco-evolutionary dynamics of host-phage interactions that are pervasive in all ecosystems.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Bondy-Denomy J, Davidson AR. When a virus is not a parasite: the beneficial effects of prophages on bacterial fitness. J Microbiol. 2014;52:235–42. - PubMed
-
- Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008;2:579–89. - PubMed
-
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–24. - PubMed
-
- Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13:641–50. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

