Reversal of glucocorticoid resistance in paediatric acute lymphoblastic leukaemia is dependent on restoring BIM expression
- PMID: 32242100
- PMCID: PMC7283241
- DOI: 10.1038/s41416-020-0824-8
Reversal of glucocorticoid resistance in paediatric acute lymphoblastic leukaemia is dependent on restoring BIM expression
Abstract
Background: Acute lymphoblastic leukaemia (ALL) is the most common paediatric malignancy. Glucocorticoids form a critical component of chemotherapy regimens and resistance to glucocorticoid therapy is predictive of poor outcome. We have previously shown that glucocorticoid resistance is associated with upregulation of the oncogene C-MYC and failure to induce the proapoptotic gene BIM.
Methods: A high-throughput screening (HTS) campaign was carried out to identify glucocorticoid sensitisers against an ALL xenograft derived from a glucocorticoid-resistant paediatric patient. Gene expression analysis was carried out using Illumina microarrays. Efficacy, messenger RNA and protein analysis were carried out by Resazurin assay, reverse transcription-PCR and immunoblotting, respectively.
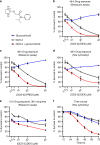
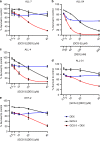
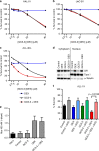
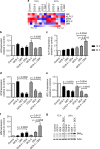
Results: A novel glucocorticoid sensitiser, 2-((4,5-dihydro-1H-imidazol-2-yl)thio)-N-isopropyl-N-phenylacetamide (GCS-3), was identified from the HTS campaign. The sensitising effect was specific to glucocorticoids and synergy was observed in a range of dexamethasone-resistant and dexamethasone-sensitive xenografts representative of B-ALL, T-ALL and Philadelphia chromosome-positive ALL. GCS-3 in combination with dexamethasone downregulated C-MYC and significantly upregulated BIM expression in a glucocorticoid-resistant ALL xenograft. The GCS-3/dexamethasone combination significantly increased binding of the glucocorticoid receptor to a novel BIM enhancer, which is associated with glucocorticoid sensitivity.
Conclusions: This study describes the potential of the novel glucocorticoid sensitiser, GCS-3, as a biological tool to interrogate glucocorticoid action and resistance.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells.Blood. 2015 Jan 8;125(2):273-83. doi: 10.1182/blood-2014-05-576470. Epub 2014 Oct 21. Blood. 2015. PMID: 25336632
-
Epigenetic silencing of BIM in glucocorticoid poor-responsive pediatric acute lymphoblastic leukemia, and its reversal by histone deacetylase inhibition.Blood. 2010 Oct 21;116(16):3013-22. doi: 10.1182/blood-2010-05-284968. Epub 2010 Jul 20. Blood. 2010. PMID: 20647567
-
Modulation of Glucocorticoid Resistance in Pediatric T-cell Acute Lymphoblastic Leukemia by Increasing BIM Expression with the PI3K/mTOR Inhibitor BEZ235.Clin Cancer Res. 2016 Feb 1;22(3):621-32. doi: 10.1158/1078-0432.CCR-15-0114. Epub 2015 Jun 16. Clin Cancer Res. 2016. PMID: 26080839 Free PMC article.
-
Glucocorticoid resistance in paediatric acute lymphoblastic leukaemia.J Paediatr Child Health. 2012 Aug;48(8):634-40. doi: 10.1111/j.1440-1754.2011.02212.x. Epub 2011 Nov 3. J Paediatr Child Health. 2012. PMID: 22050419 Review.
-
[Association between BIM gene and glucocorticoid resistance in children with acute lymphoblastic leukemia].Zhongguo Dang Dai Er Ke Za Zhi. 2017 Aug;19(8):945-949. doi: 10.7499/j.issn.1008-8830.2017.08.018. Zhongguo Dang Dai Er Ke Za Zhi. 2017. PMID: 28774373 Free PMC article. Review. Chinese.
Cited by
-
Overcoming Glucocorticoid Resistance in Acute Lymphoblastic Leukemia: Repurposed Drugs Can Improve the Protocol.Front Oncol. 2021 Mar 11;11:617937. doi: 10.3389/fonc.2021.617937. eCollection 2021. Front Oncol. 2021. PMID: 33777761 Free PMC article. Review.
-
JUN mediates glucocorticoid resistance by stabilizing HIF1a in T cell acute lymphoblastic leukemia.iScience. 2023 Oct 18;26(11):108242. doi: 10.1016/j.isci.2023.108242. eCollection 2023 Nov 17. iScience. 2023. PMID: 38026210 Free PMC article.
-
Salidroside overcomes dexamethasone resistance in T-acute lymphoblastic leukemia cells.Exp Ther Med. 2021 Jun;21(6):636. doi: 10.3892/etm.2021.10068. Epub 2021 Apr 15. Exp Ther Med. 2021. PMID: 33968167 Free PMC article.
-
Dasatinib overcomes glucocorticoid resistance in B-cell acute lymphoblastic leukemia.Nat Commun. 2023 May 22;14(1):2935. doi: 10.1038/s41467-023-38456-y. Nat Commun. 2023. PMID: 37217509 Free PMC article.
-
Precursor B-ALL Cell Lines Differentially Respond to SYK Inhibition by Entospletinib.Int J Mol Sci. 2021 Jan 8;22(2):592. doi: 10.3390/ijms22020592. Int J Mol Sci. 2021. PMID: 33435587 Free PMC article.
References
-
- Youlden DR, Gupta S, Frazier AL, Moore AS, Baade PD, Valery PC, et al. Stage at diagnosis for children with blood cancers in Australia: application of the Toronto Paediatric Cancer Stage Guidelines in a population-based national childhood cancer registry. Pediatr. Blood Cancer. 2019;66:e27683. - PubMed
-
- Bhojwani D, Pui C-H. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14:205–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

