Enhancing the sensitivity of the thymidine kinase assay by using DNA repair-deficient human TK6 cells
- PMID: 32243652
- PMCID: PMC7384079
- DOI: 10.1002/em.22371
Enhancing the sensitivity of the thymidine kinase assay by using DNA repair-deficient human TK6 cells
Abstract
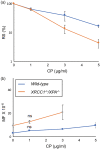
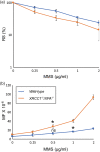
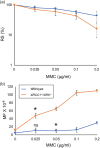
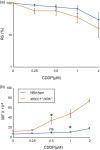
The OECD guidelines define the bioassays of identifying mutagenic chemicals, including the thymidine kinase (TK) assay, which specifically detects the mutations that inactivate the TK gene in the human TK6 lymphoid line. However, the sensitivity of this assay is limited because it detects mutations occurring only in the TK gene but not any other genes. Moreover, the limited sensitivity of the conventional TK assay is caused by the usage of DNA repair-proficient wild-type cells, which are capable of accurately repairing DNA damage induced by chemicals. Mutagenic chemicals produce a variety of DNA lesions, including base lesions, sugar damage, crosslinks, and strand breaks. Base damage causes point mutations and is repaired by the base excision repair (BER) and nucleotide excision repair (NER) pathways. To increase the sensitivity of TK assay, we simultaneously disrupted two genes encoding XRCC1, an important BER factor, and XPA, which is essential for NER, generating XRCC1 -/- /XPA -/- cells from TK6 cells. We measured the mutation frequency induced by four typical mutagenic agents, methyl methane sulfonate (MMS), cis-diamminedichloro-platinum(II) (cisplatin, CDDP), mitomycin-C (MMC), and cyclophosphamide (CP) by the conventional TK assay using wild-type TK6 cells and also by the TK assay using XRCC1 -/- /XPA -/- cells. The usage of XRCC1 -/- /XPA -/- cells increased the sensitivity of detecting the mutagenicity by 8.6 times for MMC, 8.5 times for CDDP, and 2.6 times for MMS in comparison with the conventional TK assay. In conclusion, the usage of XRCC1 -/- /XPA -/- cells will significantly improve TK assay.
Keywords: TK assay; DNA-damaging agent; OECD guideline; TK6 cells; thymidine kinase assay.
© 2020 The Authors. Environmental and Molecular Mutagenesis published by Wiley Periodicals, Inc. on behalf of Environmental Mutagen Society.
Figures
Similar articles
-
Follow-up genotoxicity assessment of Ames-positive/equivocal chemicals using the improved thymidine kinase gene mutation assay in DNA repair-deficient human TK6 cells.Mutagenesis. 2021 Oct 6;36(5):331-338. doi: 10.1093/mutage/geab025. Mutagenesis. 2021. PMID: 34216473
-
Differential micronucleus frequency in isogenic human cells deficient in DNA repair pathways is a valuable indicator for evaluating genotoxic agents and their genotoxic mechanisms.Environ Mol Mutagen. 2018 Jul;59(6):529-538. doi: 10.1002/em.22201. Epub 2018 May 15. Environ Mol Mutagen. 2018. PMID: 29761828
-
An assay to detect DNA-damaging agents that induce nucleotide excision-repairable DNA lesions in living human cells.Mutat Res Genet Toxicol Environ Mutagen. 2017 Aug;820:1-7. doi: 10.1016/j.mrgentox.2017.05.009. Epub 2017 May 22. Mutat Res Genet Toxicol Environ Mutagen. 2017. PMID: 28676261
-
Xeroderma Pigmentosa Group A (XPA), Nucleotide Excision Repair and Regulation by ATR in Response to Ultraviolet Irradiation.Adv Exp Med Biol. 2017;996:41-54. doi: 10.1007/978-3-319-56017-5_4. Adv Exp Med Biol. 2017. PMID: 29124689 Free PMC article. Review.
-
DNA repair-deficient Xpa and Xpa/p53+/- knock-out mice: nature of the models.Toxicol Pathol. 2001;29 Suppl:109-16. doi: 10.1080/019262301753178519. Toxicol Pathol. 2001. PMID: 11695546 Review.
Cited by
-
Comprehensive whole-genome sequencing reveals origins of mutational signatures associated with aging, mismatch repair deficiency and temozolomide chemotherapy.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1122. doi: 10.1093/nar/gkae1122. Nucleic Acids Res. 2025. PMID: 39656916 Free PMC article.
-
Dual-directional epi-genotoxicity assay for assessing chemically induced epigenetic effects utilizing the housekeeping TK gene.Sci Rep. 2025 Mar 5;15(1):7780. doi: 10.1038/s41598-025-92121-6. Sci Rep. 2025. PMID: 40044744 Free PMC article.
-
REV7-p53 interaction inhibits ATM-mediated DNA damage signaling.Cell Cycle. 2024 Feb;23(4):339-352. doi: 10.1080/15384101.2024.2333227. Epub 2024 Apr 1. Cell Cycle. 2024. PMID: 38557443 Free PMC article.
References
-
- Aboussekhra, A. , Biggerstaff, M. , Shivji, M.K.K. , Vilpo, J.A. , Moncollin, V. , Podust, V.N. , Protić, M. , Hübscher, U. , Egly, J.‐M. and Wood, R.D. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. - PubMed
-
- Beranek, D.T. (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutation Research, 231, 11–30. - PubMed
-
- Caldecott, K.W. (2003) DNA single‐strand break repair and spinocerebellar ataxia. Cell, 112, 7–10. - PubMed
-
- Cermak, T. , Doyle, E.L. , Christian, M. , Wang, L. , Zhang, Y. , Schmidt, C. , Baller, J.A. , Somia, N.V. , Bogdanove, A.J. and Voytas, D.F. (2011) Efficient design and assembly of custom TALEN and other TAL effector‐based constructs for DNA targeting. Nucleic Acids Research, 39, e82–e82. - PMC - PubMed
-
- Clements, J. (1995) Gene mutation assays in mammalian cells. Methods in Molecular Biology, 43, 277–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

