Human iPSC-Derived Neurons and Cerebral Organoids Establish Differential Effects of Germline NF1 Gene Mutations
- PMID: 32243842
- PMCID: PMC7160375
- DOI: 10.1016/j.stemcr.2020.03.007
Human iPSC-Derived Neurons and Cerebral Organoids Establish Differential Effects of Germline NF1 Gene Mutations
Abstract
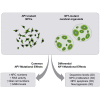
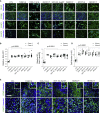
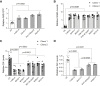
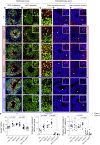
Neurofibromatosis type 1 (NF1) is a common neurodevelopmental disorder caused by a spectrum of distinct germline NF1 gene mutations, traditionally viewed as equivalent loss-of-function alleles. To specifically address the issue of mutational equivalency in a disease with considerable clinical heterogeneity, we engineered seven isogenic human induced pluripotent stem cell lines, each with a different NF1 patient NF1 mutation, to identify potential differential effects of NF1 mutations on human central nervous system cells and tissues. Although all mutations increased proliferation and RAS activity in 2D neural progenitor cells (NPCs) and astrocytes, we observed striking differences between NF1 mutations on 2D NPC dopamine levels, and 3D NPC proliferation, apoptosis, and neuronal differentiation in developing cerebral organoids. Together, these findings demonstrate differential effects of NF1 gene mutations at the cellular and tissue levels, suggesting that the germline NF1 gene mutation is one factor that underlies clinical variability.
Keywords: disease modeling; human iPSCs; neurodevelopment; neurofibromatosis type 1.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Bolcekova A., Nemethova M., Zatkova A., Hlinkova K., Pozgayova S., Hlavata A., Kadasi L., Durovcikova D., Gerinec A., Husakova K. Clustering of mutations in the 5' tertile of the NF1 gene in Slovakia patients with optic pathway glioma. Neoplasma. 2013;60:655–665. - PubMed
-
- Costa R.M., Federov N.B., Kogan J.H., Murphy G.G., Stern J., Ohno M., Kucherlapati R., Jacks T., Silva A.J. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

