DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes
- PMID: 32243864
- PMCID: PMC7212265
- DOI: 10.1016/j.ajhg.2020.03.008
DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes
Abstract
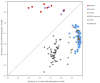
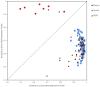
Weaver syndrome (WS), an overgrowth/intellectual disability syndrome (OGID), is caused by pathogenic variants in the histone methyltransferase EZH2, which encodes a core component of the Polycomb repressive complex-2 (PRC2). Using genome-wide DNA methylation (DNAm) data for 187 individuals with OGID and 969 control subjects, we show that pathogenic variants in EZH2 generate a highly specific and sensitive DNAm signature reflecting the phenotype of WS. This signature can be used to distinguish loss-of-function from gain-of-function missense variants and to detect somatic mosaicism. We also show that the signature can accurately classify sequence variants in EED and SUZ12, which encode two other core components of PRC2, and predict the presence of pathogenic variants in undiagnosed individuals with OGID. The discovery of a functionally relevant signature with utility for diagnostic classification of sequence variants in EZH2, EED, and SUZ12 supports the emerging paradigm shift for implementation of DNAm signatures into diagnostics and translational research.
Keywords: DNA methylation signature; EED; SUZ12; intellectual disability; overgrowth syndromes.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Tatton-Brown K., Murray A., Hanks S., Douglas J., Armstrong R., Banka S., Bird L.M., Clericuzio C.L., Cormier-Daire V., Cushing T., Childhood Overgrowth Consortium Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am. J. Med. Genet. A. 2013;161A:2972–2980. - PubMed
-
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. - PubMed
-
- Cohen A.S., Tuysuz B., Shen Y., Bhalla S.K., Jones S.J., Gibson W.T. A novel mutation in EED associated with overgrowth. J. Hum. Genet. 2015;60:339–342. - PubMed

