The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation
- PMID: 32244383
- PMCID: PMC7232374
- DOI: 10.3390/v12040384
The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation
Abstract
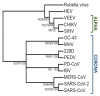
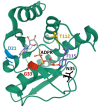
Macrodomains, enzymes that remove ADP-ribose from proteins, are encoded by several families of RNA viruses and have recently been shown to counter innate immune responses to virus infection. ADP-ribose is covalently attached to target proteins by poly-ADP-ribose polymerases (PARPs), using nicotinamide adenine dinucleotide (NAD+) as a substrate. This modification can have a wide variety of effects on proteins including alteration of enzyme activity, protein-protein interactions, and protein stability. Several PARPs are induced by interferon (IFN) and are known to have antiviral properties, implicating ADP-ribosylation in the host defense response and suggesting that viral macrodomains may counter this response. Recent studies have demonstrated that viral macrodomains do counter the innate immune response by interfering with PARP-mediated antiviral defenses, stress granule formation, and pro-inflammatory cytokine production. Here, we will describe the known functions of the viral macrodomains and review recent literature demonstrating their roles in countering PARP-mediated antiviral responses.
Keywords: ADP-ribose; ADP-ribosylation; PARPs; RNA virus; alphaviruses; coronaviruses; hepatitis E virus; macrodomain; stress granule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Koonin E.V., Gorbalenya A.E., Purdy M.A., Rozanov M.N., Reyes G.R., Bradley D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. - DOI - PMC - PubMed
-
- Gorbalenya A.E., Koonin E.V., Lai M.M.-C. Putative papain-related thiol proteases of positive-strand RNA viruses Identification of rubi-and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, α-and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. - DOI - PMC - PubMed
-
- Malet H., Coutard B., Jamal S., Dutartre H., Papageorgiou N., Neuvonen M., Ahola T., Forrester N., Gould E.A., Lafitte D., et al. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. - DOI - PMC - PubMed
-
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

