Human Melanoma-Cell Metabolic Profiling: Identification of Novel Biomarkers Indicating Metastasis
- PMID: 32244549
- PMCID: PMC7177954
- DOI: 10.3390/ijms21072436
Human Melanoma-Cell Metabolic Profiling: Identification of Novel Biomarkers Indicating Metastasis
Abstract
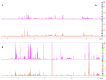
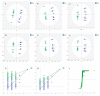
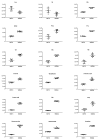
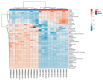
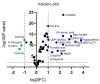
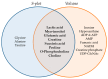
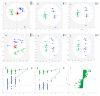
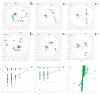
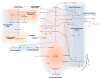
Melanoma is the most aggressive type of skin cancer, leading to metabolic rewiring and enhancement of metastatic transformation. Efforts to improve its early and accurate diagnosis are largely based on preclinical models and especially cell lines. Hence, we herein present a combinational Nuclear Magnetic Resonance (NMR)- and Ultra High Performance Liquid Chromatography-High-Resolution Tandem Mass Spectrometry (UHPLC-HRMS/MS)-mediated untargeted metabolomic profiling of melanoma cells, to landscape metabolic alterations likely controlling metastasis. The cell lines WM115 and WM2664, which belong to the same patient, were examined, with WM115 being derived from a primary, pre-metastatic, tumor and WM2664 clonally expanded from lymph-node metastases. Metabolite samples were analyzed using NMR and UHPLC-HRMS. Multivariate statistical analysis of high resolution NMR and MS (positive and negative ionization) results was performed by Principal Component Analysis (PCA), Partial Least Squares-Discriminant Analysis (PLS-DA) and Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA), while metastasis-related biomarkers were determined on the basis of VIP lists, S-plots and Student's t-tests. Receiver Operating Characteristic (ROC) curves of NMR and MS data revealed significantly differentiated metabolite profiles for each cell line, with WM115 being mainly characterized by upregulated levels of phosphocholine, choline, guanosine and inosine. Interestingly, WM2664 showed notably increased contents of hypoxanthine, myo-inositol, glutamic acid, organic acids, purines, pyrimidines, AMP, ADP, ATP and UDP(s), thus indicating the critical roles of purine, pyrimidine and amino acid metabolism during human melanoma metastasis.
Keywords: MS; NMR; biomarker; cancer; melanoma; metabolomics; metastasis.
Conflict of interest statement
The authors declare that have no competing financial and/or non-financial interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

