Bacterial Factors Targeting the Nucleus: The Growing Family of Nucleomodulins
- PMID: 32244550
- PMCID: PMC7232420
- DOI: 10.3390/toxins12040220
Bacterial Factors Targeting the Nucleus: The Growing Family of Nucleomodulins
Abstract
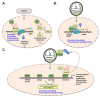
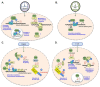
Pathogenic bacteria secrete a variety of proteins that manipulate host cell function by targeting components of the plasma membrane, cytosol, or organelles. In the last decade, several studies identified bacterial factors acting within the nucleus on gene expression or other nuclear processes, which has led to the emergence of a new family of effectors called "nucleomodulins". In human and animal pathogens, Listeria monocytogenes for Gram-positive bacteria and Anaplasma phagocytophilum, Ehrlichia chaffeensis, Chlamydia trachomatis,Legionella pneumophila, Shigella flexneri, and Escherichia coli for Gram-negative bacteria, have led to pioneering discoveries. In this review, we present these paradigms and detail various mechanisms and core elements (e.g., DNA, histones, epigenetic regulators, transcription or splicing factors, signaling proteins) targeted by nucleomodulins. We particularly focus on nucleomodulins interacting with epifactors, such as LntA of Listeria and ankyrin repeat- or tandem repeat-containing effectors of Rickettsiales, and nucleomodulins from various bacterial species acting as post-translational modification enzymes. The study of bacterial nucleomodulins not only generates important knowledge about the control of host responses by microbes but also creates new tools to decipher the dynamic regulations that occur in the nucleus. This research also has potential applications in the field of biotechnology. Finally, this raises questions about the epigenetic effects of infectious diseases.
Keywords: Listeria; effectors; epigenetics; nucleomodulin; nucleus; pathogens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Van Montagu M., Holsters M., Zambryski P., Hernalsteens J.P., Depicker A., De Beuckeleer M., Engler G., Lemmers M., Willmitzer L., Schell J. The interaction of Agrobacterium Ti-plasmid DNA and plant cells. Proc. R. Soc. Lond. B Biol. Sci. 1980;210:351–365. doi: 10.1098/rspb.1980.0139. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

