The Treasure Vault Can be Opened: Large-Scale Genome Skimming Works Well Using Herbarium and Silica Gel Dried Material
- PMID: 32244605
- PMCID: PMC7238428
- DOI: 10.3390/plants9040432
The Treasure Vault Can be Opened: Large-Scale Genome Skimming Works Well Using Herbarium and Silica Gel Dried Material
Abstract
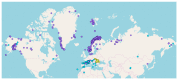
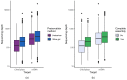
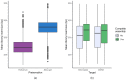
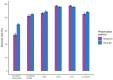
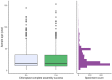
Genome skimming has the potential for generating large data sets for DNA barcoding and wider biodiversity genomic studies, particularly via the assembly and annotation of full chloroplast (cpDNA) and nuclear ribosomal DNA (nrDNA) sequences. We compare the success of genome skims of 2051 herbarium specimens from Norway/Polar regions with 4604 freshly collected, silica gel dried specimens mainly from the European Alps and the Carpathians. Overall, we were able to assemble the full chloroplast genome for 67% of the samples and the full nrDNA cluster for 86%. Average insert length, cover and full cpDNA and rDNA assembly were considerably higher for silica gel dried than herbarium-preserved material. However, complete plastid genomes were still assembled for 54% of herbarium samples compared to 70% of silica dried samples. Moreover, there was comparable recovery of coding genes from both tissue sources (121 for silica gel dried and 118 for herbarium material) and only minor differences in assembly success of standard barcodes between silica dried (89% ITS2, 96% matK and rbcL) and herbarium material (87% ITS2, 98% matK and rbcL). The success rate was > 90% for all three markers in 1034 of 1036 genera in 160 families, and only Boraginaceae worked poorly, with 7 genera failing. Our study shows that large-scale genome skims are feasible and work well across most of the land plant families and genera we tested, independently of material type. It is therefore an efficient method for increasing the availability of plant biodiversity genomic data to support a multitude of downstream applications.
Keywords: ITS; alpine; chloroplast DNA; environmental DNA; matK; nuclear ribosomal DNA; phylogenomic; plant DNA barcode; polar; rbcL.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Grants and funding
- 226134/F50/Norges Forskningsråd
- 14-14, 70184209/Norwegian Biodiversity Information Centre
- PhD internal funded/Universitetet i Tromsø
- FP7/2007-2013 281422/Seventh Framework Programme
- GOCE-CT-2007-036866/Sixth Framework Programme
- Grant 31003A_149508/1/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung
- NextBarcode project/Institut Français de Bioinformatique
- ANR-07-BDIV-014/Agence Nationale de la Recherche
- ANR-13-ISV7-0004/Agence Nationale de la Recherche
- PN-II-ID-JRP-RO-FR-2012/Agence Nationale de la Recherche
- BRANCUSI No. 32660WB/Agence Nationale de la Recherche
- PN-II-CT-ROFR-2014-2-0011/Agence Nationale de la Recherche
- PN-II-RU-PD-2012-3-0636/Romanian Ministry of Education
- ANR-10-INBS-09-08/France Génomique
LinkOut - more resources
Full Text Sources
Miscellaneous

