The Interplay between Mucosal Microbiota Composition and Host Gene-Expression is Linked with Infliximab Response in Inflammatory Bowel Diseases
- PMID: 32244928
- PMCID: PMC7143962
- DOI: 10.3390/microorganisms8030438
The Interplay between Mucosal Microbiota Composition and Host Gene-Expression is Linked with Infliximab Response in Inflammatory Bowel Diseases
Abstract
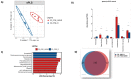
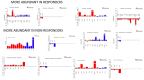
Even though anti-TNF therapy significantly improves the rates of remission in inflammatory bowel disease (IBD) patients, there is a noticeable subgroup of patients who do not respond to treatment. Dysbiosis emerges as a key factor in IBD pathogenesis. The aim of the present study is to profile changes in the gut microbiome and transcriptome before and after administration of the anti-TNF agent Infliximab (IFX) and investigate their potential to predict patient response to IFX at baseline. Mucosal biopsy samples from 20 IBD patients and nine healthy controls (HC) were examined for differences in microbiota composition (16S rRNA gene sequencing) and mucosal gene expression (RT-qPCR) at baseline and upon completion of IFX treatment, accordingly, via an in silico pipeline. Significant differences in microbiota composition were found between the IBD and HC groups. Several bacterial genera, which were found only in IBD patients and not HC, had their populations dramatically reduced after anti-TNF treatment regardless of response. Alpha and beta diversity metrics showed significant differences between our study groups. Correlation analysis revealed six microbial genera associated with differential expression of inflammation-associated genes in IFX treatment responders at baseline. This study shows that IFX treatment has a notable impact on both the gut microbial composition and the inflamed tissue transcriptome in IBD patients. Importantly, our results identify enterotypes that correlate with transcriptome changes and help differentiate IFX responders versus non-responders at baseline, suggesting that, in combination, these signatures can be an effective tool to predict anti-TNF response.
Keywords: anti-TNF; biomarkers; host transcriptome; inflammatory bowel disease; infliximab; microbiome; microbiota; response to therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

