Computational Study on the Effect of Inactivating/Activating Mutations on the Inhibition of MEK1 by Trametinib
- PMID: 32245216
- PMCID: PMC7139317
- DOI: 10.3390/ijms21062167
Computational Study on the Effect of Inactivating/Activating Mutations on the Inhibition of MEK1 by Trametinib
Abstract
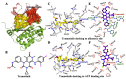
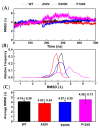
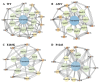
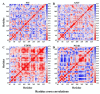
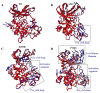
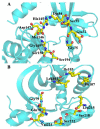
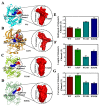
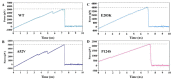
Activation of the mitogen-activated protein kinase (MAPK) signaling pathway regulated by human MAP kinase 1 (MEK1) is associated with the carcinogenesis and progression of numerous cancers. In addition, two active mutations (P124S and E203K) have been reported to enhance the activity of MEK1, thereby eventually leading to the tumorigenesis of cancer. Trametinib is an MEK1 inhibitor for treating EML4-ALK-positive, EGFR-activated, and KRAS-mutant lung cancers. Therefore, in this study, molecular docking and molecular dynamic (MD) simulations were performed to explore the effects of inactive/active mutations (A52V/P124S and E203K) on the conformational changes of MEK1 and the changes in the interaction of MEK1 with trametinib. Moreover, steered molecular dynamic (SMD) simulations were further utilized to compare the dissociation processes of trametinib from the wild-type (WT) MEK1 and two active mutants (P124S and E203K). As a result, trametinib had stronger interactions with the non-active MEK1 (WT and A52V mutant) than the two active mutants (P124S and E203K). Moreover, two active mutants may make the allosteric channel of MEK1 wider and shorter than that of the non-active types (WT and A52V mutant). Hence, trametinib could dissociate from the active mutants (P124S and E203K) more easily compared with the WT MEK1. In summary, our theoretical results demonstrated that the active mutations may attenuate the inhibitory effects of MEK inhibitor (trametinib) on MEK1, which could be crucial clues for future anti-cancer treatment.
Keywords: MEK1; allosteric channel; docking; molecular dynamic simulations; steered molecular dynamic simulations; trametinib.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Study of Molecular Mechanism of the Interaction Between MEK1/2 and Trametinib with Docking and Molecular Dynamic Simulation.Interdiscip Sci. 2019 Mar;11(1):115-124. doi: 10.1007/s12539-018-0305-4. Epub 2018 Nov 21. Interdiscip Sci. 2019. PMID: 30465279
-
Trametinib in the treatment of multiple malignancies harboring MEK1 mutations.Cancer Treat Rev. 2019 Dec;81:101907. doi: 10.1016/j.ctrv.2019.101907. Epub 2019 Oct 14. Cancer Treat Rev. 2019. PMID: 31715422 Review.
-
Allosteric MEK1/2 inhibitors including cobimetanib and trametinib in the treatment of cutaneous melanomas.Pharmacol Res. 2017 Mar;117:20-31. doi: 10.1016/j.phrs.2016.12.009. Epub 2016 Dec 9. Pharmacol Res. 2017. PMID: 27956260 Review.
-
Effects of F53 hotspot mutations on the molecular dynamics of MEK1 protein and the binding of its FDA-approved inhibitors.Int J Biol Macromol. 2025 May;306(Pt 1):141329. doi: 10.1016/j.ijbiomac.2025.141329. Epub 2025 Feb 20. Int J Biol Macromol. 2025. PMID: 39986525
-
MEK inhibitor trametinib does not prevent the growth of anaplastic lymphoma kinase (ALK)-addicted neuroblastomas.Sci Signal. 2017 Nov 28;10(507):eaam7550. doi: 10.1126/scisignal.aam7550. Sci Signal. 2017. PMID: 29184034
Cited by
-
Molecular and Pathological Profiling of Corresponding Treatment-Naïve and Neoadjuvant Pazopanib-Treated High-Risk Soft Tissue Sarcoma Samples of the GISG-04/NOPASS Study.Biology (Basel). 2021 Jul 9;10(7):639. doi: 10.3390/biology10070639. Biology (Basel). 2021. PMID: 34356494 Free PMC article.
-
Molecular Dockings and Molecular Dynamics Simulations Reveal the Potency of Different Inhibitors against Xanthine Oxidase.ACS Omega. 2021 Apr 22;6(17):11639-11649. doi: 10.1021/acsomega.1c00968. eCollection 2021 May 4. ACS Omega. 2021. PMID: 34056319 Free PMC article.
-
Investigating the Retained Inhibitory Effect of Cobimetinib against p.P124L Mutated MEK1: A Combined Liquid Biopsy and in Silico Approach.Cancers (Basel). 2022 Aug 27;14(17):4153. doi: 10.3390/cancers14174153. Cancers (Basel). 2022. PMID: 36077693 Free PMC article.
-
Uncovering the effects and molecular mechanism of Astragalus membranaceus (Fisch.) Bunge and its bioactive ingredients formononetin and calycosin against colon cancer: An integrated approach based on network pharmacology analysis coupled with experimental validation and molecular docking.Front Pharmacol. 2023 Jan 23;14:1111912. doi: 10.3389/fphar.2023.1111912. eCollection 2023. Front Pharmacol. 2023. PMID: 36755950 Free PMC article.
-
Conformational Changes of Glutamine 5'-Phosphoribosylpyrophosphate Amidotransferase for Two Substrates Analogue Binding: Insight from Conventional Molecular Dynamics and Accelerated Molecular Dynamics Simulations.Front Chem. 2021 Feb 26;9:640994. doi: 10.3389/fchem.2021.640994. eCollection 2021. Front Chem. 2021. PMID: 33718330 Free PMC article.
References
-
- Zheng C.F., Guan K.L. Cloning and characterization of two distinct human extracellular signal-regulated kinase activator kinases, MEK1 and MEK2. J. Biol. Chem. 1993;268:11435–11439. - PubMed
-
- Robarge K.D., Lee W., Eigenbrot C., Ultsch M., Wiesmann C., Heald R., Price S., Hewitt J., Jackson P., Savy P., et al. Structure based design of novel 6,5 heterobicyclic mitogen-activated protein kinase kinase (MEK) inhibitors leading to the discovery of imidazo[1,5-a] pyrazine G-479. Bioorg. Med. Chem. Lett. 2014;24:4714–4723. doi: 10.1016/j.bmcl.2014.08.008. - DOI - PubMed
-
- Ohren J.F., Chen H.F., Pavlovsky A., Whitehead C., Zhang E.L., Kuffa P., Yan C.H., McConnell P., Spessard C., Banotai C., et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

