1 H-Imidazole-2,5-Dicarboxamides as NS4A Peptidomimetics: Identification of a New Approach to Inhibit HCV-NS3 Protease
- PMID: 32245218
- PMCID: PMC7175367
- DOI: 10.3390/biom10030479
1 H-Imidazole-2,5-Dicarboxamides as NS4A Peptidomimetics: Identification of a New Approach to Inhibit HCV-NS3 Protease
Abstract
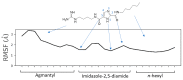
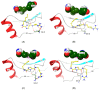
The nonstructural (NS) protein NS3/4A protease is a critical factor for hepatitis C virus (HCV) maturation that requires activation by NS4A. Synthetic peptide mutants of NS4A were found to inhibit NS3 function. The bridging from peptide inhibitors to heterocyclic peptidomimetics of NS4A has not been considered in the literature and, therefore, we decided to explore this strategy for developing a new class of NS3 inhibitors. In this report, a structure-based design approach was used to convert the bound form of NS4A into 1H-imidazole-2,5-dicarboxamide derivatives as first generation peptidomimetics. This scaffold mimics the buried amino acid sequence Ile-25` to Arg-28` at the core of NS4A21`-33` needed to activate the NS3 protease. Some of the synthesized compounds (Coded MOC) were able to compete with and displace NS4A21`-33` for binding to NS3. For instance, N5-(4-guanidinobutyl)-N2-(n-hexyl)-1H-imidazole-2,5-dicarboxamide (MOC-24) inhibited the binding of NS4A21`-33` with a competition half maximal inhibitory concentration (IC50) of 1.9 ± 0.12 µM in a fluorescence anisotropy assay and stabilized the denaturation of NS3 by increasing the aggregation temperature (40% compared to NS4A21`-33`). MOC-24 also inhibited NS3 protease activity in a fluorometric assay. Molecular dynamics simulations were conducted to rationalize the differences in structure-activity relationship (SAR) between the active MOC-24 and the inactive MOC-26. Our data show that MOC compounds are possibly the first examples of NS4A peptidomimetics that have demonstrated promising activities against NS3 proteins.
Keywords: DSLS; Flaviviridae; NS3 inhibitors; NS4A; allosteric inhibitors; binding assay; hepatitis C virus; imidazole; molecular dynamics; peptidomimetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- WHO Global Hepatitis Report. [(accessed on 21 January 2020)];2017 Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
-
- Friedrich M.J. Third millennium challenge: Hepatitis C. JAMA. 1999;282:221–222. - PubMed
-
- Stanaway J.D., Flaxman A.D., Naghavi M., Fitzmaurice C., Vos T., Abubakar I., Abu-Raddad L.J., Assadi R., Bhala N., Cowie B., et al. The global burden of viral hepatitis from 1990 to 2013: Findings from the Global Burden of Disease Study 2013. Lancet. 2016;388:1081–1088. doi: 10.1016/S0140-6736(16)30579-7. - DOI - PMC - PubMed
-
- North C.S., Hong B.A., Adewuyi S.A., Pollio D.E., Jain M.K., Devereaux R., Quartey N.A., Ashitey S., Lee W.M., Lisker-Melman M. Hepatitis C treatment and SVR: The gap between clinical trials and real-world treatment aspirations. Gen. Hosp. Psychiatry. 2013;35:122–128. doi: 10.1016/j.genhosppsych.2012.11.002. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

