Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria
- PMID: 32245284
- PMCID: PMC7143992
- DOI: 10.3390/microorganisms8030447
Identification and in vitro Characterization of a Novel Phage Endolysin that Targets Gram-Negative Bacteria
Abstract
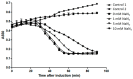
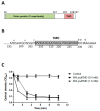
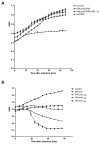
Most double-stranded (ds) DNA phages utilize holin proteins to secrete endolysin for host peptidoglycan lysis. In contrast, several holin-independent endolysins with secretion sequences or signal-arrest-release (SAR) sequences are secreted via the Sec pathway. In this study, we characterized a novel lysis protein (M4Lys) encoded by the dsDNA phage BSPM4, whose lysis function is not dependent on either holin or the Sec pathway in vitro. In silico analysis of M4Lys revealed that it contains a putative virion protein domain and an unusual C-terminal transmembrane domain (TMD). Turbidity reduction assays and liquid chromatography-mass spectrometry using purified peptidoglycan showed that the virion protein domain of M4Lys has peptidoglycan lysis activity. In vitro overproduction of M4Lys in Escherichia coli revealed that M4Lys alone caused rapid cell lysis. Treatment of E. coli with a Sec inhibitor did not inhibit the lysis activity of M4Lys, indicating that the Sec pathway is not involved in M4Lys-mediated cell lysis. Truncation of the TMD eliminated the cell lysis phenomenon, while production of the TMD alone did not induce the cell lysis. All these findings demonstrate that M4Lys is a novel endolysin that has a unique mosaic structure distinct from other canonical endolysins and the TMD plays a critical role in M4Lys-mediated in vitro cell lysis.
Keywords: cell lysis kinetics; endolysin; flagella-targeting phage; secretion; transmembrane domain.
Conflict of interest statement
The authors have no competing financial interests to declare.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

