Genome-wide analysis of differentially expressed profiles of mRNAs, lncRNAs and circRNAs in chickens during Eimeria necatrix infection
- PMID: 32245514
- PMCID: PMC7118956
- DOI: 10.1186/s13071-020-04047-9
Genome-wide analysis of differentially expressed profiles of mRNAs, lncRNAs and circRNAs in chickens during Eimeria necatrix infection
Abstract
Background: Eimeria necatrix, the most highly pathogenic coccidian in chicken small intestines, can cause high morbidity and mortality in susceptible birds and devastating economic losses in poultry production, but the underlying molecular mechanisms in interaction between chicken and E. necatrix are not entirely revealed. Accumulating evidence shows that the long-non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) are key regulators in various infectious diseases. However, the expression profiles and roles of these two non-coding RNAs (ncRNAs) during E. necatrix infection are still unclear.
Methods: The expression profiles of mRNAs, lncRNAs and circRNAs in mid-segments of chicken small intestines at 108 h post-infection (pi) with E. necatrix were analyzed by using the RNA-seq technique.
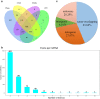
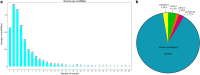
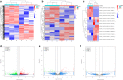
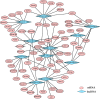
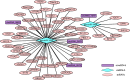
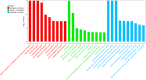
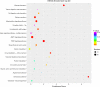
Results: After strict filtering of raw data, we putatively identified 49,183 mRNAs, 818 lncRNAs and 4153 circRNAs. The obtained lncRNAs were classified into four types, including 228 (27.87%) intergenic, 67 (8.19%) intronic, 166 (20.29%) anti-sense and 357 (43.64%) sense-overlapping lncRNAs; of these, 571 were found to be novel. Five types were also predicted for putative circRNAs, including 180 exonic, 54 intronic, 113 antisense, 109 intergenic and 3697 sense-overlapping circRNAs. Eimeria necatrix infection significantly altered the expression of 1543 mRNAs (707 upregulated and 836 downregulated), 95 lncRNAs (49 upregulated and 46 downregulated) and 13 circRNAs (9 upregulated and 4 downregulated). Target predictions revealed that 38 aberrantly expressed lncRNAs would cis-regulate 73 mRNAs, and 1453 mRNAs could be trans-regulated by 87 differentially regulated lncRNAs. Additionally, 109 potential sponging miRNAs were also identified for 9 circRNAs. GO and KEGG enrichment analysis of target mRNAs for lncRNAs, and sponging miRNA targets and source genes for circRNAs identified associations of both lncRNAs and circRNAs with host immune defense and pathogenesis during E. necatrix infection.
Conclusions: To the best of our knowledge, the present study provides the first genome-wide analysis of mRNAs, lncRNAs and circRNAs in chicken small intestines infected with E. necatrix. The obtained data will offer novel clues for exploring the interaction mechanisms between chickens and Eimeria spp.
Keywords: Chicken small intestine; Eimeria necatrix; Expression profile; circRNAs; lncRNAs; mRNAs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
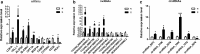
Similar articles
-
Transcriptome analysis of differentially expressed circRNAs miRNAs and mRNAs during the challenge of coccidiosis.Front Immunol. 2022 Nov 15;13:910860. doi: 10.3389/fimmu.2022.910860. eCollection 2022. Front Immunol. 2022. PMID: 36458003 Free PMC article.
-
Micro-RNA expression profile of chicken small intestines during Eimeria necatrix infection.Poult Sci. 2020 May;99(5):2444-2451. doi: 10.1016/j.psj.2019.12.065. Epub 2020 Mar 11. Poult Sci. 2020. PMID: 32359579 Free PMC article.
-
Genome-wide analysis of differentially expressed profiles of mRNAs, lncRNAs and circRNAs during Cryptosporidium baileyi infection.BMC Genomics. 2018 May 10;19(1):356. doi: 10.1186/s12864-018-4754-2. BMC Genomics. 2018. PMID: 29747577 Free PMC article.
-
CircRNAs and LncRNAs in Osteoporosis.Differentiation. 2020 Nov-Dec;116:16-25. doi: 10.1016/j.diff.2020.10.002. Epub 2020 Oct 28. Differentiation. 2020. PMID: 33157509 Review.
-
Roles of lncRNAs and circRNAs in regulating skeletal muscle development.Acta Physiol (Oxf). 2020 Feb;228(2):e13356. doi: 10.1111/apha.13356. Epub 2019 Aug 19. Acta Physiol (Oxf). 2020. PMID: 31365949 Review.
Cited by
-
Full-length transcriptome analysis and identification of transcript structures in Eimeria necatrix from different developmental stages by single-molecule real-time sequencing.Parasit Vectors. 2021 Sep 27;14(1):502. doi: 10.1186/s13071-021-05015-7. Parasit Vectors. 2021. PMID: 34579769 Free PMC article.
-
Comprehensive Analyses of circRNA Expression Profiles and Function Prediction in Chicken Cecums After Eimeria tenella Infection.Front Cell Infect Microbiol. 2021 Mar 10;11:628667. doi: 10.3389/fcimb.2021.628667. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33777841 Free PMC article.
-
Emerging roles of circular RNAs on the regulation of production traits in chicken.Poult Sci. 2025 Jan;104(1):104612. doi: 10.1016/j.psj.2024.104612. Epub 2024 Nov 28. Poult Sci. 2025. PMID: 39647355 Free PMC article. Review.
-
Transcriptome analysis of differentially expressed circRNAs miRNAs and mRNAs during the challenge of coccidiosis.Front Immunol. 2022 Nov 15;13:910860. doi: 10.3389/fimmu.2022.910860. eCollection 2022. Front Immunol. 2022. PMID: 36458003 Free PMC article.
-
RNA sequencing reveals dynamic expression of lncRNAs and mRNAs in caprine endometrial epithelial cells induced by Neospora caninum infection.Parasit Vectors. 2022 Aug 24;15(1):297. doi: 10.1186/s13071-022-05405-5. Parasit Vectors. 2022. PMID: 35999576 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

