A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV
- PMID: 32245784
- PMCID: PMC7164391
- DOI: 10.1126/science.abb7269
A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV
Abstract
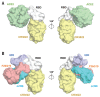
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) has now become a pandemic, but there is currently very little understanding of the antigenicity of the virus. We therefore determined the crystal structure of CR3022, a neutralizing antibody previously isolated from a convalescent SARS patient, in complex with the receptor binding domain (RBD) of the SARS-CoV-2 spike (S) protein at 3.1-angstrom resolution. CR3022 targets a highly conserved epitope, distal from the receptor binding site, that enables cross-reactive binding between SARS-CoV-2 and SARS-CoV. Structural modeling further demonstrates that the binding epitope can only be accessed by CR3022 when at least two RBDs on the trimeric S protein are in the "up" conformation and slightly rotated. These results provide molecular insights into antibody recognition of SARS-CoV-2.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures


References
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). 10.1016/S0140-6736(20)30183-5 - DOI - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
-
- ter Meulen J., van den Brink E. N., Poon L. L. M., Marissen W. E., Leung C. S. W., Cox F., Cheung C. Y., Bakker A. Q., Bogaards J. A., van Deventer E., Preiser W., Doerr H. W., Chow V. T., de Kruif J., Peiris J. S. M., Goudsmit J., Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLOS Med. 3, e237 (2006). 10.1371/journal.pmed.0030237 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

