Defective nucleotide-dependent assembly and membrane fusion in Mfn2 CMT2A variants improved by Bax
- PMID: 32245838
- PMCID: PMC7136618
- DOI: 10.26508/lsa.201900527
Defective nucleotide-dependent assembly and membrane fusion in Mfn2 CMT2A variants improved by Bax
Abstract
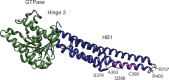
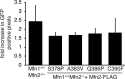
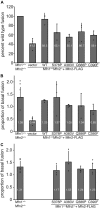
Mitofusins are members of the dynamin-related protein family of large GTPases that harness the energy from nucleotide hydrolysis to remodel membranes. Mitofusins possess four structural domains, including a GTPase domain, two extended helical bundles (HB1 and HB2), and a transmembrane region. We have characterized four Charcot-Marie-Tooth type 2A-associated variants with amino acid substitutions in Mfn2 that are proximal to the hinge that connects HB1 and HB2. A functional defect was not apparent in cells as the mitochondrial morphology of Mfn2-null cells was restored by expression of any of these variants. However, a significant fusion deficiency was observed in vitro, which was improved by the addition of crude cytosol extract or soluble Bax. All four variants had reduced nucleotide-dependent assembly in cis, but not trans, and this was also improved by the addition of Bax. Together, our data demonstrate an important role for this region in Mfn2 GTP-dependent oligomerization and membrane fusion and is consistent with a model where cytosolic factors such as Bax are masking molecular defects associated with Mfn2 disease variants in cells.
© 2020 Samanas et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures









References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
